Isolation and Functional Analysis of ZmLTP3, a Homologue to Arabidopsis LTP3
Abstract
:1. Introduction
2. Results
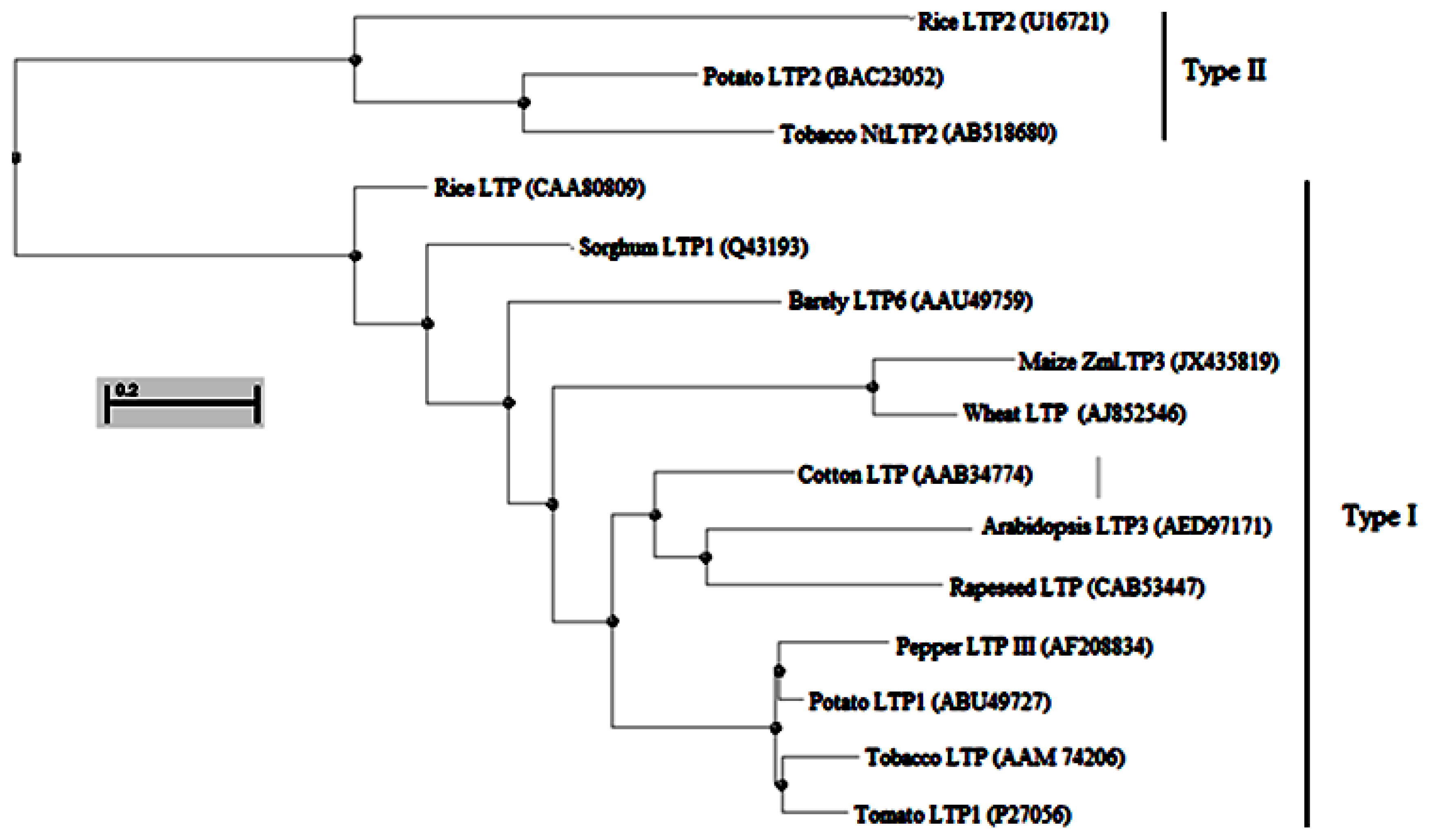
2.1. Isolation and Sequence Analysis of ZmLTP3
2.2. Expression Analysis of ZmLTP3
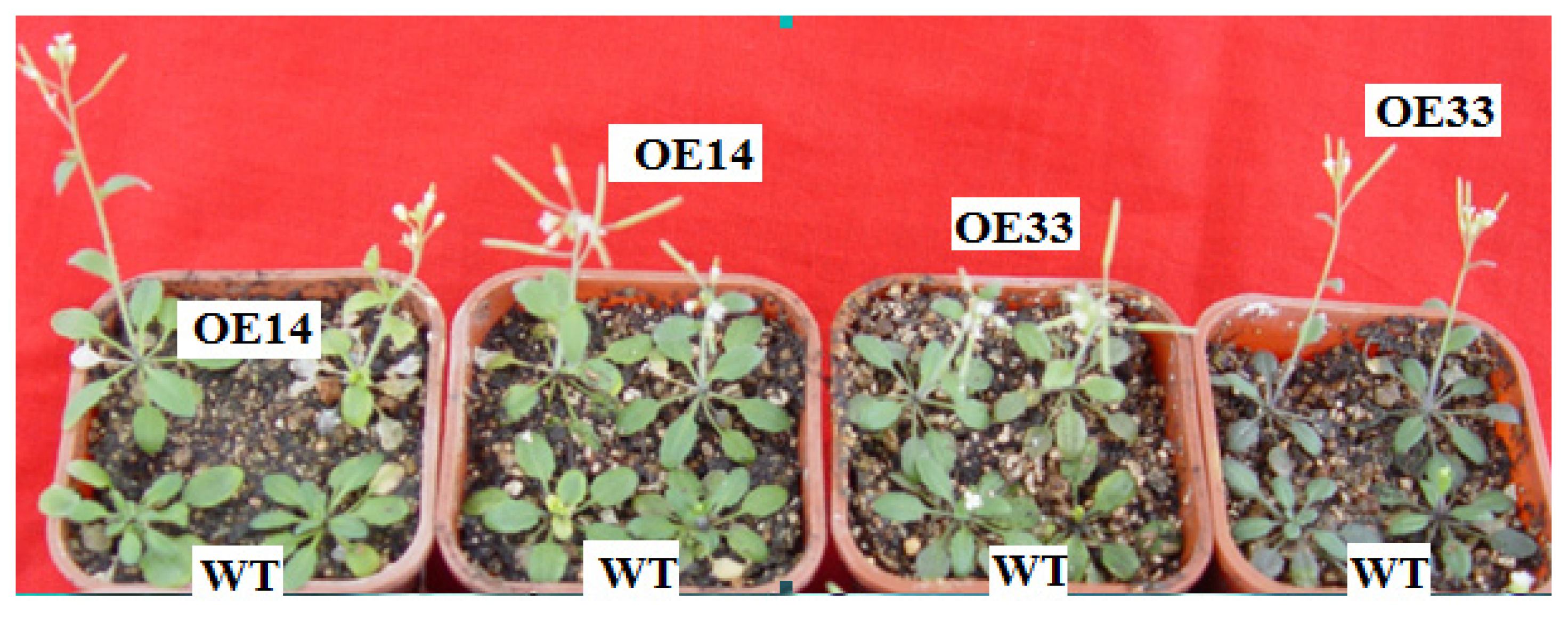
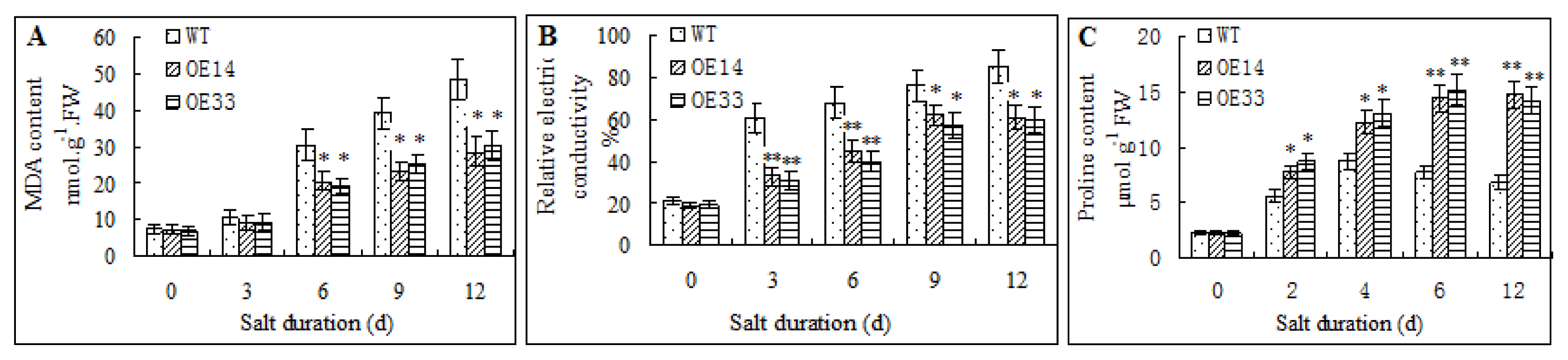
2.3. Salt Tolerance of Transgenic Arabidopsis
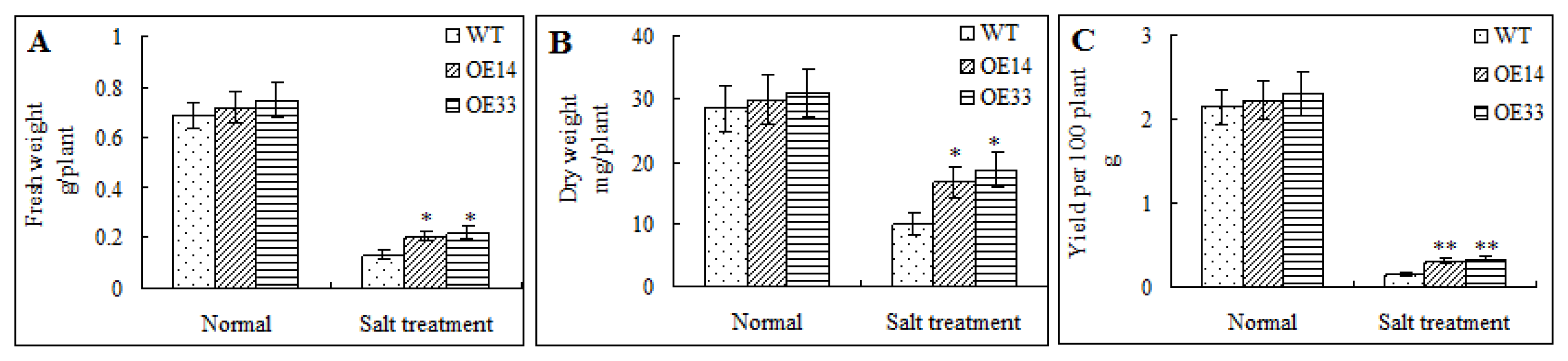
2.4. Morphological and Physiological Analysis of Transgenic Arabidopsis
3. Discussion
4. Experimental Section
4.1. Maize Seedlings Growth Conditions and Treatments
4.2. Cloning of ZmLTP3 cDNA
4.3. RT-PCR Analysis of ZmLTP3
4.4. Plasimid Construction, Arabidopsis Transformation and Transformants Characterization
4.5. Arabidopsis Growth Conditions and Stress Treatments
4.6. Morphological and Physiological Traits Measurements
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Bernhard, W.R.; Thoma, S.; Botella, J.; Somerville, C.R. Isolation of a cDNA clone for spinach lipid transfer protein and evidence that the protein is synthesized by the secretory pathway. Plant Physiol 1991, 95, 164–170. [Google Scholar]
- Kader, J.C. Lipid transfer proteins in plants. Annu. Rev. Plant Physiol. Mol. Biol 1996, 47, 627–654. [Google Scholar]
- Thoma, S.; Hecht, U.; Kippers, A.; Botella, J.; Devries, S.; Somerville, C. Tissue specific expression of a gene encoding a cell wall localized lipid transfer protein from Arabidopsis. Plant Physiol 1994, 105, 35–45. [Google Scholar]
- Carvalho, A.O.; Gomes, V.M. Role of plant lipid transfer proteins in plant cell physiology: A concise review. Peptides 2007, 28, 1144–1153. [Google Scholar]
- Douliez, J.P.; Michon, T.; Elmorjani, K.; Marion, D. Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J. Cereal Sci 2000, 32, 1–20. [Google Scholar]
- Zachowski, A.; Guerbette, F.; Grosbois, M. Characterization of acyl binding by a plant lipid-transfer protein. Eur. J. Biochem 1998, 257, 443–448. [Google Scholar]
- Kinlaw, C.S.; Gerttula, S.M.; Carter, M.C. Lipid transfer protein genes of loblolly pine are members of a complex gene family. Plant Mol. Biol 1994, 26, 1213–1216. [Google Scholar]
- Feng, J.X.; Ji, S.J.; Shi, Y.H.; Wei, G.; Zhu, Y.X. Analysis of five differentially expressed gene families in fast elongating cotton fiber. Acta Biochim. Biophys. Sin 2004, 36, 51–57. [Google Scholar]
- Liu, K.; Jiang, H.; Moore, S.; Watkins, C.; Jahn, M. Isolation and characterization of a lipid transfer protein expressed in ripening fruit of Capsicum chinense. Planta 2006, 223, 672–683. [Google Scholar]
- Boutrot, F.; Meynard, D.; Guiderdoni, E.; Joudrier, P.; Gautier, M.F. The Triticum aestivum non-specific lipid transfer protein (TaLtp) gene family: Comparative promoter activity of six TaLtp genes in transgenic rice. Planta 2007, 225, 843–862. [Google Scholar]
- Boutrot, F.; Chantret, N.; Gautier, M.F. Genome-wide analysis of the rice and Arabidopsis non-specific lipid transfer protein (nsLtp) gene families and identification of wheat nsLtp genes by EST data mining. BMC Genomics 2008, 9, 86–105. [Google Scholar]
- Choi, Y.E.; Lim, S.; Kim, H.J.; Han, J.Y.; Lee, M.H.; Yang, Y.; Kim, J.A.; Kim, Y.S. Tobacco NtLTP1, a glandular-specific lipid transfer protein, is required for lipid secretion from glandular trichomes. Plant J 2012, 70, 480–491. [Google Scholar]
- Tsuboi, S.; Osafune, T.; Tsugeki, R.; Nishimura, M.; Yamada, M. Nonspecific lipid transfer protein in castor bean cotyledon cells: Subcellular localization and a possible role in lipid metabolism. J. Biochem 1992, 111, 500–508. [Google Scholar]
- Hollenbach, B.; Schreiber, L.; Hartung, W.; Dietz, K.J. Cadmium leads to stimulated expression of the lipid transfer protein genes in barley: Implications for the involvement of lipid transfer proteins in wax assembly. Planta 1997, 203, 9–19. [Google Scholar]
- Park, S.Y.; Lord, E.M. Expression studies of SCA in lily and confirmation of its role in pollen tube adhesion. Plant Mol. Biol 2003, 51, 183–189. [Google Scholar]
- Yeats, T.H.; Rose, J.K.C. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sic 2008, 17, 191–198. [Google Scholar]
- Lee, S.B.; Go, Y.S.; Bae, H.J.; Park, J.H.; Cho, S.H.; Cho, H.J.; Lee, D.S.; Park, O.K.; Hwang, I.; Suh, M.C. Disruption of glycosylphosphatidylinositolanchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiol 2009, 150, 42–54. [Google Scholar]
- Sarowar, S.; Kim, Y.J.; Kim, K.D.; Hwang, B.K.; Ok, S.H.; Shin, J.S. Overexpression of lipid transfer protein (LTP) genes enhances resistance to plant pathogens and LTP functions in long-distance systemic signaling in tobacco. Plant Cell Rep 2009, 28, 419–427. [Google Scholar]
- Van Loon, L.C.; van Strien, E.A. The families of pathogenesis related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol 1999, 55, 85–97. [Google Scholar]
- Zou, H.W.; Wu, Z.Y.; Yang, Q.; Zhang, X.H.; Cao, M.Q.; Jia, W.S.; Huang, C.L.; Xiao, X. Gene expression analyses of ZmPti1, encoding a maize Pti-like kinase, suggest a role in stress signaling. Plant Sci 2006, 171, 99–105. [Google Scholar]
- Zou, H.W.; Wu, Z.Y.; Zhang, X.H.; Wang, Y.Q.; Huang, C.L. Over-expression of ZmPti1, a homologue to Pti1, increases salt tolerance of Arabidopsis thaliana. Afr. J. Biotech 2010, 9, 656–662. [Google Scholar]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 2003, 31, 3784–3788. [Google Scholar]
- Benson, D.A.; Karsch-Mizrachi, I.; Clark, K.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res 2012, 40, D48–D53. [Google Scholar]
- Mittova, V.; Guy, M.; Ta, M.; Volokita, M. Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii. J. Exp. Bot 2004, 55, 1105–1113. [Google Scholar]
- Armengaud, P.; Thiery, L.; Buhot, N.; Grenier-De, M.G.; Savoure, A. Transcriptional regulation of proline biosynthesis in Medicago truncatula reveals developmental and environmental specific features. Physiol. Plant 2004, 120, 442–450. [Google Scholar]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplast: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophysi 1968, 125, 189–198. [Google Scholar]



© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zou, H.-W.; Tian, X.-H.; Ma, G.-H.; Li, Z.-X. Isolation and Functional Analysis of ZmLTP3, a Homologue to Arabidopsis LTP3. Int. J. Mol. Sci. 2013, 14, 5025-5035. https://doi.org/10.3390/ijms14035025
Zou H-W, Tian X-H, Ma G-H, Li Z-X. Isolation and Functional Analysis of ZmLTP3, a Homologue to Arabidopsis LTP3. International Journal of Molecular Sciences. 2013; 14(3):5025-5035. https://doi.org/10.3390/ijms14035025
Chicago/Turabian StyleZou, Hua-Wen, Xiao-Hai Tian, Guo-Hui Ma, and Zhi-Xin Li. 2013. "Isolation and Functional Analysis of ZmLTP3, a Homologue to Arabidopsis LTP3" International Journal of Molecular Sciences 14, no. 3: 5025-5035. https://doi.org/10.3390/ijms14035025



