Cold Signaling and Cold Response in Plants
Abstract
:1. Introduction
2. Cold Stress Sensing and Second Messengers
3. ICE-CBF/DREB1 Pathway and Cold-Responsive Gene Regulation
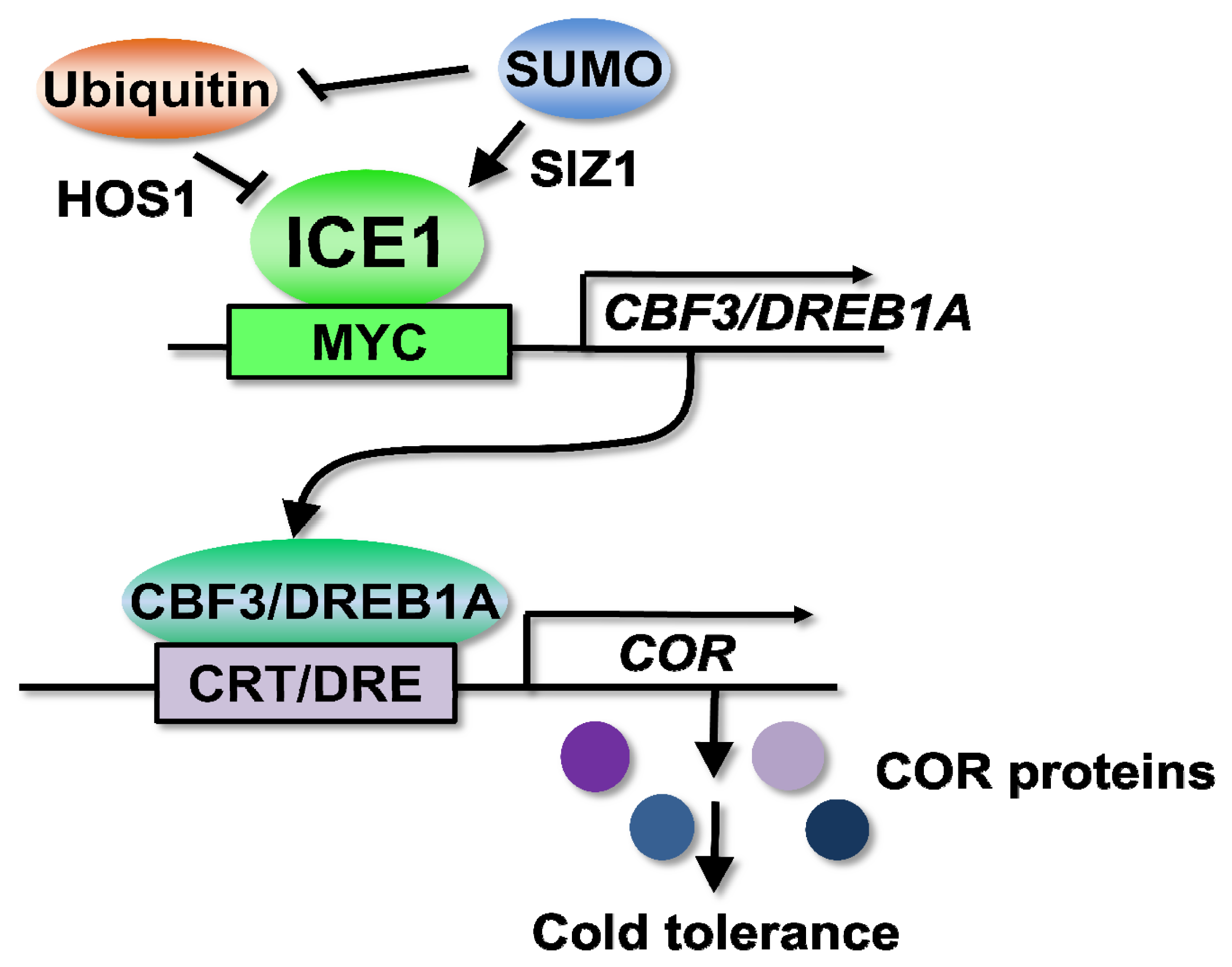
3.1. ICEs (Inducer of CBF Expressions) Are Transcription Factors Controlling Cold Signaling through the Regulation of CBF/DREB1s
3.2. The CBF/DREB1 Responsive Pathway
3.3. Cold-Regulated Genes
4. Post-Transcriptional Regulation
5. Post-Translational Regulation
6. Cold Response and Plant Hormones
7. Chloroplast and Cold Response
8. Conclusions and Perspective
Acknowledgements
References
- Ramankutty, N.; Evan, A.T.; Monfreda, C.; Foley, J.A. Farming the planet: 1. Geographic distribution of global agricultural lands in the year 2000. Glob. Biogeochem. Cycles 2008, 22. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol 1999, 50, 571–599. [Google Scholar]
- Chinnusamy, V.; Zhu, J.K.; Sunkar, R. Gene regulation during cold stress acclimation in plants. Methods Mol. Biol 2010, 639, 39–55. [Google Scholar]
- Hannah, M.A.; Heyer, A.G.; Hincha, D.K. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 2005, 1, e26. [Google Scholar]
- Orvar, B.L.; Sangwan, V.; Omann, F.; Dhindsa, R.S. Early steps in cold sensing by plant cells: The role of actin cytoskeleton and membrane fluidity. Plant J 2000, 23, 785–794. [Google Scholar]
- Sangwan, V.; Foulds, I.; Singh, J.; Dhindsa, R.S. Cold-Activation of Brassica napus BN115 promoter is mediated by structural changes in membranes and cytoskeleton, and requires Ca2+ influx. Plant J 2001, 27, 1–12. [Google Scholar]
- Knight, M.R.; Campbell, A.K.; Smith, S.M.; Trewavas, A.J. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 1991, 352, 524–526. [Google Scholar]
- Van der Luit, A.H.; Olivari, C.; Haley, A.; Knight, M.R.; Trewavas, A.J. Distinct calcium signaling pathways regulate calmodulin gene expression in tobacco. Plant Physiol 1999, 121, 705–714. [Google Scholar]
- Mauger, J.P. Role of the nuclear envelope in calcium signalling. Biol. Cell 2012, 104, 70–83. [Google Scholar]
- Xu, X.M.; Meier, I. The nuclear pore comes to the fore. Trends Plant Sci 2008, 13, 20–27. [Google Scholar]
- Mazars, C.; Brière, C.; Bourque, S.; Thuleau, P. Nuclear calcium signaling: An emerging topic in plants. Biochimie 2011, 93, 2068–2074. [Google Scholar]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 2000, 23, 319–327. [Google Scholar]
- Townley, H.E.; Knight, M.R. Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol 2002, 128, 1169–1172. [Google Scholar]
- Huang, C.; Ding, S.; Zhang, H.; Du, H.; An, L. CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Sci 2011, 181, 57–64. [Google Scholar]
- Doherty, C.J.; van Buskirk, H.A.; Myers, S.J.; Thomashow, M.F. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 2009, 21, 972–984. [Google Scholar]
- Teige, M.; Scheikl, E.; Eulgem, T.; Doczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar]
- Sangwan, V.; Orvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J 2002, 31, 629–638. [Google Scholar]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci 2007, 12, 444–451. [Google Scholar]
- Gilmour, S.J.; Fowler, S.G.; Thomashow, M.F. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol 2004, 54, 767–781. [Google Scholar]
- Gilmour, S.J.; Sebolt, A.M.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 2000, 124, 1854–1865. [Google Scholar]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 2003, 17, 1043–1054. [Google Scholar]
- Lee, B.H.; Henderson, D.A.; Zhu, J.K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 2005, 17, 3155–3175. [Google Scholar]
- Fursova, O.V.; Pogorelko, G.V.; Tarasov, V.A. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 2009, 429, 98–103. [Google Scholar]
- Hu, Y.; Zhang, L.; Zhao, L.; Li, J.; He, S.; Zhou, K.; Yang, F.; Huang, M.; Jiang, L.; Li, L. Trichostatin A selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS One 2011, 6, e22132. [Google Scholar]
- Dong, C.H.; Agarwal, M.; Zhang, Y.; Xie, Q.; Zhu, J.K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 2006, 103, 8281–8286. [Google Scholar]
- Miura, K.; Jin, J.B.; Lee, J.; Yoo, C.Y.; Stirm, V.; Miura, T.; Ashworth, E.N.; Bressan, R.A.; Yun, D.J.; Hasegawa, P.M. SIZ1-Mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 2007, 19, 1403–1414. [Google Scholar]
- Miura, K.; Shiba, H.; Ohta, M.; Kang, S.W.; Sato, A.; Yuasa, T.; Iwaya-Inoue, M.; Kamada, H.; Ezura, H. SlICE1 encoding a MYC-type transcription factor controls cold tolerance in tomato, Solanum lycopersicum. Plant Biotechnol 2012, 29, 253–260. [Google Scholar]
- Miura, K.; Sato, A.; Shiba, H.; Kang, S.W.; Kamada, H.; Ezura, H. Accumulation of antioxidants and antioxidant activity in tomato, Solanum lycopersicum, are enhanced by the transcription factor SlICE1. Plant Biotechnol 2012, 29, 261–269. [Google Scholar]
- Badawi, M.; Reddy, Y.V.; Agharbaoui, Z.; Tominaga, Y.; Danyluk, J.; Sarhan, F.; Houde, M. Structure and functional analysis of wheat ICE (inducer of CBF expression) genes. Plant Cell Physiol 2008, 49, 1237–1249. [Google Scholar]
- Nakamura, J.; Yuasa, T.; Huong, T.T.; Harano, K.; Tanaka, S.; Iwata, T.; Phan, T.; Iwaya-Inoue, M. Rice homologs of inducer of CBF expression (OsICE) are involved in cold acclimation. Plant Biotechnol 2011, 28, 303–309. [Google Scholar]
- Zhao, M.L.; Wang, J.N.; Shan, W.; Fan, J.G.; Kuang, J.F.; Wu, K.Q.; Li, X.P.; Chen, W.X.; He, F.Y.; Chen, J.Y.; et al. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ 2012, 36, 30–51. [Google Scholar]
- Liu, L.; Duan, L.; Zhang, J.; Zhang, Z.; Mi, G.; Ren, H. Cucumber (Cucumis sativus L.) over-expressing cold-induced transcriptome regulator ICE1 exhibits changed morphological characters and enhances chilling tolerance. Sci. Hortic 2010, 124, 29–33. [Google Scholar]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar]
- Ito, Y.; Katsura, K.; Maruyama, K.; Taji, T.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 2006, 47, 141–153. [Google Scholar]
- Oh, S.J.; Kwon, C.W.; Choi, D.W.; Song, S.I.; Kim, J.K. Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol. J 2007, 5, 646–656. [Google Scholar]
- Morran, S.; Eini, O.; Pyvovarenko, T.; Parent, B.; Singh, R.; Ismagul, A.; Eliby, S.; Shirley, N.; Langridge, P.; Lopato, S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J 2011, 9, 230–249. [Google Scholar]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 2001, 127, 910–917. [Google Scholar]
- Owens, C.L.; Thomashow, M.F.; Hancock, J.F.; Iezzoni, A.F. CBF1 orthologs in sour cherry and strawberry and the heterologous expression of CBF1 in strawberry. J. Am. Soc. Hortic. Sci 2002, 127, 489–494. [Google Scholar]
- Behnam, B.; Kikuchi, A.; Celebi-Toprak, F.; Kasuga, M.; Yamaguchi-Shinozaki, K.; Watanabe, K. Arabidopsis rd29A::DREB1A enhances freezing tolerance in transgenic potato. Plant Cell. Rep 2007, 26, 1275–1282. [Google Scholar]
- Benedict, C.; Skinner, J.S.; Meng, R.; Chang, Y.; Bhalerao, R.; Huner, N.P.; Finn, C.E.; Chen, T.H.; Hurry, V. The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ 2006, 29, 1259–1272. [Google Scholar]
- Pellegrineschi, A.; Reynolds, M.; Pacheco, M.; Brito, R.M.; Almeraya, R.; Yamaguchi-Shinozaki, K.; Hoisington, D. Stress-Induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 2004, 47, 493–500. [Google Scholar]
- Kasuga, M.; Miura, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol 2004, 45, 346–350. [Google Scholar]
- Zhang, X.; Fowler, S.G.; Cheng, H.; Lou, Y.; Rhee, S.Y.; Stockinger, E.J.; Thomashow, M.F. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J 2004, 39, 905–919. [Google Scholar]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 2003, 33, 751–763. [Google Scholar]
- Qin, F.; Sakuma, Y.; Li, J.; Liu, Q.; Li, Y.Q.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol 2004, 45, 1042–1052. [Google Scholar]
- Siddiqua, M.; Nassuth, A. Vitis CBF1 and Vitis CBF4 differ in their effect on Arabidopsis abiotic stress tolerance, development and gene expression. Plant Cell Environ 2011, 34, 1345–1359. [Google Scholar]
- Skinner, J.S.; von Zitzewitz, J.; Szucs, P.; Marquez-Cedillo, L.; Filichkin, T.; Amundsen, K.; Stockinger, E.J.; Thomashow, M.F.; Chen, T.H.; Hayes, P.M. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol. Biol 2005, 59, 533–551. [Google Scholar]
- Xiong, Y.; Fei, S.-Z. Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 2006, 224, 878–888. [Google Scholar]
- Zhao, H.; Bughrara, S.S. Isolation and characterization of cold-regulated transcriptional activator LpCBF3 gene from perennial ryegrass (Lolium perenne L.). Mol. Genet. Genomics 2008, 279, 585–594. [Google Scholar]
- Hsieh, T.H.; Lee, J.T.; Yang, P.T.; Chiu, L.H.; Charng, Y.Y.; Wang, Y.C.; Chan, M.T. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol 2002, 129, 1086–1094. [Google Scholar]
- Yang, W.; Liu, X.D.; Chi, X.J.; Wu, C.A.; Li, Y.Z.; Song, L.L.; Liu, X.M.; Wang, Y.F.; Wang, F.W.; Zhang, C.; Liu, Y.; et al. Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 2011, 233, 219–229. [Google Scholar]
- Chen, M.; Xu, Z.; Xia, L.; Li, L.; Cheng, X.; Dong, J.; Wang, Q.; Ma, Y. Cold-Induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). J. Exp. Bot 2009, 60, 121–135. [Google Scholar]
- Welling, A.; Palva, E.T. Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol 2008, 147, 1199–1211. [Google Scholar]
- Gutha, L.; Reddy, A. Rice DREB1B promoter shows distinct stress-specific responses, and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol. Biol 2008, 68, 533–555. [Google Scholar]
- Maruyama, K.; Sakuma, Y.; Kasuga, M.; Ito, Y.; Seki, M.; Goda, H.; Shimada, Y.; Yoshida, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 2004, 38, 982–993. [Google Scholar]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 2012, 1819, 86–96. [Google Scholar]
- Medina, J.; Bargues, M.; Terol, J.; Perez-Alonso, M.; Salinas, J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 1999, 119, 463–470. [Google Scholar]
- Medina, J.; Catalá, R.; Salinas, J. The CBFs: Three arabidopsis transcription factors to cold acclimate. Plant Sci 2011, 180, 3–11. [Google Scholar]
- Matsui, A.; Ishida, J.; Morosawa, T.; Okamoto, M.; Kim, J.M.; Kurihara, Y.; Kawashima, M.; Tanaka, M.; To, T.K.; Nakaminami, K.; et al. Arabidopsis tiling array analysis to identify the stress-responsive genes. Methods Mol. Biol 2010, 639, 141–155. [Google Scholar]
- Zeller, G.; Henz, S.R.; Widmer, C.K.; Sachsenberg, T.; Ratsch, G.; Weigel, D.; Laubinger, S. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 2009, 58, 1068–1082. [Google Scholar]
- Fowler, S.; Thomashow, M.F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002, 14, 1675–1690. [Google Scholar]
- Vogel, J.T.; Zarka, D.G.; van Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 2005, 41, 195–211. [Google Scholar]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar]
- Lee, S.C.; Huh, K.W.; An, K.; An, G.; Kim, S.R. Ectopic expression of a cold-inducible transcription factor, CBF1/DREB1b, in transgenic rice (Oryza sativa L.). Mol. Cells 2004, 18, 107–114. [Google Scholar]
- Francia, E.; Barabaschi, D.; Tondelli, A.; Laido, G.; Rizza, F.; Stanca, A.M.; Busconi, M.; Fogher, C.; Stockinger, E.J.; Pecchioni, N. Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor. Appl. Genet 2007, 115, 1083–1091. [Google Scholar]
- Stockinger, E.J.; Skinner, J.S.; Gardner, K.G.; Francia, E.; Pecchioni, N. Expression levels of barley Cbf genes at the Frost resistance-H2 locus are dependent upon alleles at Fr-H1 and Fr-H2. Plant J 2007, 51, 308–321. [Google Scholar]
- Knox, A.K.; Li, C.; Vagujfalvi, A.; Galiba, G.; Stockinger, E.J.; Dubcovsky, J. Identification of candidate CBF genes for the frost tolerance locus Fr-Am2 in Triticum monococcum. Plant Mol. Biol 2008, 67, 257–270. [Google Scholar]
- Miller, A.K.; Galiba, G.; Dubcovsky, J. A cluster of 11 CBF transcription factors is located at the frost tolerance locus Fr-Am2 in Triticum monococcum. Mol. Genet. Genomics 2006, 275, 193–203. [Google Scholar]
- Alm, V.; Busso, C.S.; Ergon, A.; Rudi, H.; Larsen, A.; Humphreys, M.W.; Rognli, O.A. QTL analyses and comparative genetic mapping of frost tolerance, winter survival and drought tolerance in meadow fescue (Festuca pratensis Huds.). Theor. Appl. Genet 2011, 123, 369–382. [Google Scholar]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant. Physiol. Plant Mol. Biol 1996, 47, 377–403. [Google Scholar]
- Thomashow, M.F. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol 1998, 118, 1–8. [Google Scholar]
- Hundertmark, M.; Hincha, D. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 2008, 9, 118. [Google Scholar]
- Kiyosue, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thaliana. Plant Cell Physiol 1994, 35, 225–231. [Google Scholar]
- Kovacs, D.; Kalmar, E.; Torok, Z.; Tompa, P. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol 2008, 147, 381–390. [Google Scholar]
- Danyluk, J.; Perron, A.; Houde, M.; Limin, A.; Fowler, B.; Benhamou, N.; Sarhan, F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 1998, 10, 623–638. [Google Scholar]
- Houde, M.; Dallaire, S.; N’Dong, D.; Sarhan, F. Overexpression of the acidic dehydrin WCOR410 improves freezing tolerance in transgenic strawberry leaves. Plant Biotechnol. J 2004, 2, 381–387. [Google Scholar]
- Bravo, L.A.; Gallardo, J.; Navarrete, A.; Olave, N.; Martínez, J.; Alberdi, M.; Close, T.J.; Corcuera, L.J. Cryoprotective activity of a cold-induced dehydrin purified from barley. Physiol. Plant 2003, 118, 262–269. [Google Scholar]
- Wisniewski, M.; Webb, R.; Balsamo, R.; Close, T.J.; Yu, X.-M.; Griffith, M. Purification, immunolocalization, cryoprotective, and antifreeze activity of PCA60: A dehydrin from peach (Prunus persica). Physiol. Plant 1999, 105, 600–608. [Google Scholar]
- Hara, M.; Terashima, S.; Kuboi, T. Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J. Plant Physiol 2001, 158, 1333–1339. [Google Scholar]
- Timperio, A.M.; Egidi, M.G.; Zolla, L. Proteomics applied on plant abiotic stresses: Role of heat shock proteins (HSP). J. Proteomics 2008, 71, 391–411. [Google Scholar]
- Renaut, J.; Hausman, J.-F.; Wisniewski, M.E. Proteomics and low-temperature studies: Bridging the gap between gene expression and metabolism. Physiol. Plant 2006, 126, 97–109. [Google Scholar]
- Seo, P.J.; Lee, A.K.; Xiang, F.; Park, C.M. Molecular and functional profiling of Arabidopsis pathogenesis-related genes: Insights into their roles in salt response of seed germination. Plant Cell Physiol 2008, 49, 334–344. [Google Scholar]
- Zhang, R.; Wang, Y.; Liu, G.; Li, H. Cloning and characterization of a pathogenesis-related gene (ThPR10) from Tamarix hispida. Acta Biol. Crac. Ser. Bot. 2010, 52, 17–25. [Google Scholar]
- Liu, J.-J.; Ekramoddoullah, A.K.M.; Yu, X. Differential expression of multiple PR10 proteins in western white pine following wounding, fungal infection and cold-hardening. Physiol. Plant 2003, 119, 544–553. [Google Scholar]
- Lee, O.R.; Pulla, R.K.; Kim, Y.J.; Balusamy, S.R.; Yang, D.C. Expression and stress tolerance of PR10 genes from Panax ginseng C. A. Meyer. Mol. Biol. Rep 2012, 39, 2365–2374. [Google Scholar]
- Pak, J.-H.; Chung, E.-S.; Shin, S.-H.; Jeon, E.-H.; Kim, M.-J.; Lee, H.-Y.; Jeung, J.-U.; Hyung, N.-I.; Lee, J.-H.; Chung, Y.-S. Enhanced fungal resistance in Arabidopsis expressing wild rice PR-3 (OgChitIVa) encoding chitinase class IV. Plant Biotechnol. Rep 2009, 3, 147–155. [Google Scholar]
- Yeh, S.; Moffatt, B.A.; Griffith, M.; Xiong, F.; Yang, D.S.C.; Wiseman, S.B.; Sarhan, F.; Danyluk, J.; Xue, Y.Q.; Hew, C.L.; et al. Chitinase genes responsive to cold encode antifreeze proteins in winter cereals. Plant Physiol 2000, 124, 1251–1264. [Google Scholar]
- Pihakaski-Maunsbach, K.; Moffatt, B.; Testillano, P.; Risueño, M.; Yeh, S.; Griffith, M.; Maunsbach, A.B. Genes encoding chitinase-antifreeze proteins are regulated by cold and expressed by all cell types in winter rye shoots. Physiol. Plant 2001, 112, 359–371. [Google Scholar]
- Takenaka, Y.; Nakano, S.; Tamoi, M.; Sakuda, S.; Fukamizo, T. Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: Chitinase inhibitor allosamidin enhances stress tolerance. Biosci. Biotechnol. Biochem 2009, 73, 1066–1071. [Google Scholar]
- Gaudet, D.A.; Laroche, A.; Frick, M.; Huel, R.; Puchalski, B. Cold induced expression of plant defensin and lipid transfer protein transcripts in winter wheat. Physiol. Plant 2003, 117, 195–205. [Google Scholar]
- Janska, A.; Marsik, P.; Zelenkova, S.; Ovesna, J. Cold stress and acclimation—What is important for metabolic adjustment? Plant Biol 2010, 12, 395–405. [Google Scholar]
- Murata, N.; Ishizaki-Nishizawa, O.; Higashi, S.; Hayashi, H.; Tasaka, Y.; Nishida, I. Genetically engineered alteration in the chilling sensitivity of plants. Nature 1992, 356, 710–713. [Google Scholar]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar]
- C, N.D.; Danyluk, J.; Wilson, K.E.; Pocock, T.; Huner, N.P.; Sarhan, F. Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analyses. Plant Physiol 2002, 129, 1368–1381. [Google Scholar]
- Artus, N.N.; Uemura, M.; Steponkus, P.L.; Gilmour, S.J.; Lin, C.; Thomashow, M.F. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. USA 1996, 93, 13404–13409. [Google Scholar]
- Puhakainen, T.; Hess, M.W.; Makela, P.; Svensson, J.; Heino, P.; Palva, E.T. Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol. Biol 2004, 54, 743–753. [Google Scholar]
- Ge, L.F.; Chao, D.Y.; Shi, M.; Zhu, M.Z.; Gao, J.P.; Lin, H.X. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008, 228, 191–201. [Google Scholar]
- Jang, I.C.; Oh, S.J.; Seo, J.S.; Choi, W.B.; Song, S.I.; Kim, C.H.; Kim, Y.S.; Seo, H.S.; Choi, Y.D.; Nahm, B.H.; et al. Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol 2003, 131, 516–524. [Google Scholar]
- Han, J.; Xiong, J.; Wang, D.; Fu, X.D. Pre-mRNA splicing: Where and when in the nucleus. Trends Cell Biol 2011, 21, 336–343. [Google Scholar]
- Ambrosone, A.; Costa, A.; Leone, A.; Grillo, S. Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci 2012, 182, 12–18. [Google Scholar]
- Kim, J.S.; Jung, H.J.; Lee, H.J.; Kim, K.A.; Goh, C.H.; Woo, Y.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 2008, 55, 455–466. [Google Scholar]
- Gong, Z.; Dong, C.H.; Lee, H.; Zhu, J.; Xiong, L.; Gong, D.; Stevenson, B.; Zhu, J.K. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 2005, 17, 256–267. [Google Scholar]
- Dong, C.H.; Hu, X.; Tang, W.; Zheng, X.; Kim, Y.S.; Lee, B.H.; Zhu, J.K. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol 2006, 26, 9533–9543. [Google Scholar]
- Filichkin, S.A.; Priest, H.D.; Givan, S.A.; Shen, R.; Bryant, D.W.; Fox, S.E.; Wong, W.K.; Mockler, T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 2010, 20, 45–58. [Google Scholar]
- Wang, B.B.; Brendel, V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 2006, 103, 7175–7180. [Google Scholar]
- Mastrangelo, A.M.; Marone, D.; Laido, G.; de Leonardis, A.M.; de Vita, P. Alternative splicing: Enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012, 185–186, 40–49. [Google Scholar]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J 2007, 49, 1091–1107. [Google Scholar]
- Dong, M.A.; Farre, E.M.; Thomashow, M.F. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7241–7246. [Google Scholar]
- Seo, P.J.; Park, M.J.; Lim, M.H.; Kim, S.G.; Lee, M.; Baldwin, I.T.; Park, C.M. A self-regulatory circuit of circadian clock-associated1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 2012, 24, 2427–2442. [Google Scholar]
- James, A.B.; Syed, N.H.; Bordage, S.; Marshall, J.; Nimmo, G.A.; Jenkins, G.I.; Herzyk, P.; Brown, J.W.; Nimmo, H.G. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 2012, 24, 961–981. [Google Scholar]
- Seo, P.J.; Kim, M.J.; Ryu, J.Y.; Jeong, E.Y.; Park, C.M. Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat. Commun 2011, 2, 303. [Google Scholar]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet 2009, 10, 94–108. [Google Scholar]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar]
- Ding, J.; Zhou, S.; Guan, J. Finding microRNA targets in plants: Current status and perspectives. Genomics Proteomics Bioinforma 2012, 10, 264–275. [Google Scholar]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci 2012, 17, 196–203. [Google Scholar]
- Zhou, X.; Wang, G.; Sutoh, K.; Zhu, J.K.; Zhang, W. Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim. Biophys. Acta 2008, 1779, 780–788. [Google Scholar]
- Chen, L.; Zhang, Y.; Ren, Y.; Xu, J.; Zhang, Z.; Wang, Y. Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem. Biophys. Res. Commun 2012, 417, 892–896. [Google Scholar]
- Zhang, J.; Xu, Y.; Huan, Q.; Chong, K. Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 2009, 10, 449. [Google Scholar]
- Lv, D.K.; Bai, X.; Li, Y.; Ding, X.D.; Ge, Y.; Cai, H.; Ji, W.; Wu, N.; Zhu, Y.M. Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 2010, 459, 39–47. [Google Scholar]
- Zhao, M.; Ding, H.; Zhu, J.K.; Zhang, F.; Li, W.X. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol 2011, 190, 906–915. [Google Scholar]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol 2012, 196, 13–28. [Google Scholar]
- Ishitani, M.; Xiong, L.; Lee, H.; Stevenson, B.; Zhu, J.K. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 1998, 10, 1151–1161. [Google Scholar]
- Lee, H.; Xiong, L.; Gong, Z.; Ishitani, M.; Stevenson, B.; Zhu, J.K. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo—Cytoplasmic partitioning. Genes Dev 2001, 15, 912–924. [Google Scholar]
- Miura, K.; Ohta, M.; Nakazawa, M.; Ono, M.; Hasegawa, P.M. ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. Plant J 2011, 67, 269–279. [Google Scholar] [Green Version]
- Lazaro, A.; Valverde, F.; Pineiro, M.; Jarillo, J.A. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 2012, 24, 982–999. [Google Scholar]
- Lee, J.H.; Kim, S.H.; Kim, J.J.; Ahn, J.H. Alternative splicing and expression analysis of high expression of osmotically responsive genes1 (HOS1) in Arabidopsis. BMB Rep 2012, 45, 515–520. [Google Scholar]
- Lee, J.H.; Kim, J.J.; Kim, S.H.; Cho, H.J.; Kim, J.; Ahn, J.H. The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in Arabidopsis thaliana. Plant Cell Physiol 2012, 53, 1802–1814. [Google Scholar]
- Jung, J.H.; Seo, P.J.; Park, C.M. The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J. Biol. Chem 2012, 287, 43277–43287. [Google Scholar]
- Yan, J.; Wang, J.; Li, Q.; Hwang, J.R.; Patterson, C.; Zhang, H. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol 2003, 132, 861–869. [Google Scholar]
- Guerra, D.; Mastrangelo, A.M.; Lopez-Torrejon, G.; Marzin, S.; Schweizer, P.; Stanca, A.M.; del Pozo, J.C.; Cattivelli, L.; Mazzucotelli, E. Identification of a protein network interacting with TdRF1, a wheat RING ubiquitin ligase with a protective role against cellular dehydration. Plant Physiol 2012, 158, 777–789. [Google Scholar]
- Miura, K.; Hasegawa, P.M. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol 2010, 20, 223–232. [Google Scholar]
- Miura, K.; Jin, J.B.; Hasegawa, P.M. Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol 2007, 10, 495–502. [Google Scholar]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol 2010, 11, 861–871. [Google Scholar]
- Castro, P.H.; Tavares, R.M.; Bejarano, E.R.; Azevedo, H. SUMO, a heavyweight player in plant abiotic stress responses. Cell. Mol. Life Sci 2012, 69, 3269–3283. [Google Scholar]
- Miura, K.; Rus, A.; Sharkhuu, A.; Yokoi, S.; Karthikeyan, A.S.; Raghothama, K.G.; Baek, D.; Koo, Y.D.; Jin, J.B.; Bressan, R.A.; et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 2005, 102, 7760–7765. [Google Scholar]
- Miura, K.; Lee, J.; Gong, Q.; Ma, S.; Jin, J.B.; Yoo, C.Y.; Miura, T.; Sato, A.; Bohnert, H.J.; Hasegawa, P.M. SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiol 2011, 155, 1000–1012. [Google Scholar]
- Park, B.S.; Song, J.T.; Seo, H.S. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat. Commun 2011, 2, 400. [Google Scholar]
- Catala, R.; Ouyang, J.; Abreu, I.A.; Hu, Y.; Seo, H.; Zhang, X.; Chua, N.H. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 2007, 19, 2952–2966. [Google Scholar]
- Miura, K.; Okamoto, H.; Okuma, E.; Shiba, H.; Kamada, H.; Hasegawa, P.M.; Murata, Y. SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J 2012, 73, 91–104. [Google Scholar]
- Yoo, C.Y.; Miura, K.; Jin, J.B.; Lee, J.; Park, H.C.; Salt, D.E.; Yun, D.J.; Bressan, R.A.; Hasegawa, P.M. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 2006, 142, 1548–1558. [Google Scholar]
- Conti, L.; Kioumourtzoglou, D.; O’Donnell, E.; Dominy, P.; Sadanandom, A. OTS1 and OTS2 SUMO proteases link plant development and survival under salt stress. Plant Signal. Behav 2009, 4, 225–227. [Google Scholar]
- Miura, K.; Sato, A.; Ohta, M.; Furukawa, J. Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2. Planta 2011, 234, 1191–1199. [Google Scholar]
- Chen, C.C.; Chen, Y.Y.; Tang, I.C.; Liang, H.M.; Lai, C.C.; Chiou, J.M.; Yeh, K.C. Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance. Plant Physiol 2011, 156, 2225–2234. [Google Scholar]
- Lee, J.; Nam, J.; Park, H.C.; Na, G.; Miura, K.; Jin, J.B.; Yoo, C.Y.; Baek, D.; Kim, D.H.; Jeong, J.C.; et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 2007, 49, 79–90. [Google Scholar]
- Van den Burg, H.A.; Kini, R.K.; Schuurink, R.C.; Takken, F.L. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 2010, 22, 1998–2016. [Google Scholar]
- Ishida, T.; Yoshimura, M.; Miura, K.; Sugimoto, K. MMS21/HPY2 and SIZ1, Two Arabidopsis SUMO E3 Ligases, Have Distinct Functions in Development. PLoS One 2012, 7, e46897. [Google Scholar]
- Miura, K.; Lee, J.; Miura, T.; Hasegawa, P.M. SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Phys 2010, 51, 103–113. [Google Scholar]
- Jin, J.B.; Jin, Y.H.; Lee, J.; Miura, K.; Yoo, C.Y.; Kim, W.Y.; van Oosten, M.; Hyun, Y.; Somers, D.E.; Lee, I.; et al. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 2008, 53, 530–540. [Google Scholar]
- Ling, Y.; Zhang, C.; Chen, T.; Hao, H.; Liu, P.; Bressan, R.A.; Hasegawa, P.M.; Jin, J.B.; Lin, J. Mutation in SUMO E3 ligase, SIZ1, disrupts the mature female gametophyte in Arabidopsis. PLoS One 2012, 7, e29470. [Google Scholar]
- Ishida, T.; Fujiwara, S.; Miura, K.; Stacey, N.; Yoshimura, M.; Schneider, K.; Adachi, S.; Minamisawa, K.; Umeda, M.; Sugimoto, K. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 2009, 21, 2284–2297. [Google Scholar]
- Miura, K.; Lee, J.; Jin, J.B.; Yoo, C.Y.; Miura, T.; Hasegawa, P.M. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 5418–5423. [Google Scholar]
- Miura, K.; Hasegawa, P.M. Sumoylation and abscisic acid signaling. Plant Signal. Behav 2009, 4, 1176–1178. [Google Scholar]
- Park, H.C.; Choi, W.; Park, H.J.; Cheong, M.S.; Koo, Y.D.; Shin, G.; Chung, W.S.; Kim, W.Y.; Kim, M.G.; Bressan, R.A.; et al. Identification and molecular properties of SUMO-binding proteins in Arabidopsis. Mol. Cells 2011, 32, 143–151. [Google Scholar]
- Miller, M.J.; Barrett-Wilt, G.A.; Hua, Z.; Vierstra, R.D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2010, 107, 16512–16517. [Google Scholar]
- Elrouby, N.; Coupland, G. Proteome-Wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc. Natl. Acad. Sci. USA 2010, 107, 17415–17420. [Google Scholar]
- Budhiraja, R.; Hermkes, R.; Muller, S.; Schmidt, J.; Colby, T.; Panigrahi, K.; Coupland, G.; Bachmair, A. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol 2009, 149, 1529–1540. [Google Scholar]
- Stockinger, E.J.; Mao, Y.; Regier, M.K.; Triezenberg, S.J.; Thomashow, M.F. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res 2001, 29, 1524–1533. [Google Scholar]
- Vlachonasios, K.E.; Thomashow, M.F.; Triezenberg, S.J. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 2003, 15, 626–638. [Google Scholar]
- Lee, B.H.; Kapoor, A.; Zhu, J.; Zhu, J.K. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 2006, 18, 1736–1749. [Google Scholar]
- Lissarre, M.; Ohta, M.; Sato, A.; Miura, K. Cold-Responsive gene regulation during cold acclimation in plants. Plant Signal. Behav 2010, 5, 948–952. [Google Scholar]
- Chen, T.H.; Gusta, L.V. Abscisic acid-induced freezing resistance in cultured plant cells. Plant Physiol 1983, 73, 71–75. [Google Scholar]
- Lang, V.; Palva, E.T. The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol 1992, 20, 951–962. [Google Scholar]
- Knight, H.; Zarka, D.G.; Okamoto, H.; Thomashow, M.F.; Knight, M.R. Abscisic acid induces CBF gene transcription and subsequent induction of cold-regulated genes via the CRT promoter element. Plant Physiol 2004, 135, 1710–1717. [Google Scholar]
- Dolferus, R.; Jacobs, M.; Peacock, W.J.; Dennis, E.S. Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol 1994, 105, 1075–1087. [Google Scholar]
- Chandler, J.; Martinez-Zapater, J.M.; Dean, C. Mutations causing defects in the biosynthesis and response to gibberellins, abscisic acid and phytochrome B do not inhibit vernalization in Arabidopsis fca-1. Planta 2000, 210, 677–682. [Google Scholar]
- Liu, J.; Gilmour, S.J.; Thomashow, M.F.; Van Nocker, S. Cold signalling associated with vernalization in Arabidopsis thaliana does not involve CBF1 or abscisic acid. Physiol. Plant 2002, 114, 125–134. [Google Scholar]
- Penfield, S. Temperature perception and signal transduction in plants. New Phytol 2008, 179, 615–628. [Google Scholar]
- Tonkinson, C.L.; Lyndon, R.F.; Arnold, G.M.; Lenton, J.R. The effects of temperature and the Rht3 dwarfing gene on growth, cell extension, and gibberellin content and responsiveness in the wheat leaf. J. Exp. Bot 1997, 48, 963–970. [Google Scholar]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar]
- Dill, A.; Thomas, S.G.; Hu, J.; Steber, C.M.; Sun, T.P. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 2004, 16, 1392–1405. [Google Scholar]
- Kosova, K.; Prasil, I.T.; Vitamvas, P.; Dobrev, P.; Motyka, V.; Flokova, K.; Novak, O.; Tureckova, V.; Rolcik, J.; Pesek, B.; et al. Complex phytohormone responses during the cold acclimation of two wheat cultivars differing in cold tolerance, winter Samanta and spring Sandra. J. Plant Physiol 2012, 169, 567–576. [Google Scholar]
- Scott, I.M.; Clarke, S.M.; Wood, J.E.; Mur, L.A. Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol 2004, 135, 1040–1049. [Google Scholar]
- Wan, S.-B.; Tian, L.; Tian, R.-R.; Pan, Q.-H.; Zhan, J.-C.; Wen, P.-F.; Chen, J.-Y.; Zhang, P.; Wang, W.; Huang, W.-D. Involvement of phospholipase D in the low temperature acclimation-induced thermotolerance in grape berry. Plant Physiol. Biochem 2009, 47, 504–510. [Google Scholar]
- Hara, M.; Furukawa, J.; Sato, A.; Mizoguchi, T.; Miura, K. Abiotic Stress Responses in Plant; Parvaiz, A., Prasad, M.N.V., Eds.; Springer: New York, NY, USA, 2012; Volume Chapter 13, pp. 235–251. [Google Scholar]
- Kang, H.M.; Saltveit, M.E. Chilling tolerance of maize, cucumber and rice seedling leaves and roots are differentially affected by salicylic acid. Physiol. Plant 2002, 115, 571–576. [Google Scholar]
- Tasgin, E.; Atici, O.; Nalbantoglu, B.; Popova, L.P. Effects of salicylic acid and cold treatments on protein levels and on the activities of antioxidant enzymes in the apoplast of winter wheat leaves. Phytochemistry 2006, 67, 710–715. [Google Scholar]
- Mora-Herrera, M.E.; López-Delgado, H.; Castillo-Morales, A.; Foyer, C.H. Salicylic acid and H2O2 function by independent pathways in the induction of freezing tolerance in potato. Physiol. Plant 2005, 125, 430–440. [Google Scholar]
- Miura, K.; Ohta, M. SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J. Plant Physiol 2010, 167, 555–560. [Google Scholar]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar]
- Masclaux-Daubresse, C.; Purdy, S.; Lemaitre, T.; Pourtau, N.; Taconnat, L.; Renou, J.-P.; Wingler, A. Genetic Variation suggests interaction between cold acclimation and metabolic regulation of leaf senescence. Plant Physiol 2007, 143, 434–446. [Google Scholar]
- Sharabi-Schwager, M.; Lers, A.; Samach, A.; Guy, C.L.; Porat, R. Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J. Exp. Bot 2010, 61, 261–273. [Google Scholar]
- Huner, N.P.A.; Öquist, G.; Sarhan, F. Energy balance and acclimation to light and cold. Trends Plant Sci 1998, 3, 224–230. [Google Scholar]
- Ivanov, A.G.; Rosso, D.; Savitch, L.V.; Stachula, P.; Rosembert, M.; Oquist, G.; Hurry, V.; Huner, N.P. Implications of alternative electron sinks in increased resistance of PSII and PSI photochemistry to high light stress in cold-acclimated Arabidopsis thaliana. Photosynth. Res 2012, 113, 191–206. [Google Scholar]
- Oquist, G.; Huner, N.P. Photosynthesis of overwintering evergreen plants. Annu. Rev. Plant Biol 2003, 54, 329–355. [Google Scholar]
- Scheller, H.V.; Haldrup, A. Photoinhibition of photosystem I. Planta 2005, 221, 5–8. [Google Scholar]
- Tyystjarvi, E. Photoinhibition of photosystem II. Int. Rev. Cell Mol. Biol 2013, 300, 243–303. [Google Scholar]
- Takahashi, S.; Murata, N. How do environmental stresses accelerate photoinhibition? Trends Plant Sci 2008, 13, 178–182. [Google Scholar]
- Ndong, C.; Danyluk, J.; Huner, N.P.; Sarhan, F. Survey of gene expression in winter rye during changes in growth temperature, irradiance or excitation pressure. Plant Mol. Biol 2001, 45, 691–703. [Google Scholar]
- Yamori, W.; Noguchi, K.; Hikosaka, K.; Terashima, I. Phenotypic plasticity in photosynthetic temperature acclimation among crop species with different cold tolerances. Plant Physiol 2010, 152, 388–399. [Google Scholar]
- Hurry, V.M.; Malmberg, G.; Gardestrom, P.; Oquist, G. Effects of a short-term shift to low temperature and of long-term cold hardening on photosynthesis and Ribulose-1,5-Bisphosphate carboxylase/oxygenase and sucrose phosphate synthase activity in leaves of winter rye (Secale cereale L.). Plant Physiol 1994, 106, 983–990. [Google Scholar]
- Strand, A.; Hurry, V.; Gustafsson, P.; Gardestrom, P. Development of Arabidopsis thaliana leaves at low temperatures releases the suppression of photosynthesis and photosynthetic gene expression despite the accumulation of soluble carbohydrates. Plant J 1997, 12, 605–614. [Google Scholar]
- Dahal, K.; Kane, K.; Gadapati, W.; Webb, E.; Savitch, L.V.; Singh, J.; Sharma, P.; Sarhan, F.; Longstaffe, F.J.; Grodzinski, B.; et al. The effects of phenotypic plasticity on photosynthetic performance in winter rye, winter wheat and Brassica napus. Physiol. Plant 2012, 144, 169–188. [Google Scholar]
- Ivanov, A.G.; Sane, P.; Hurry, V.; Król, M.; Sveshnikov, D.; Huner, N.P.A.; Öquist, G. Low-temperature modulation of the redox properties of the acceptor side of photosystem II: photoprotection through reaction centre quenching of excess energy. Physiol. Plant 2003, 119, 376–383. [Google Scholar]
- Ivanov, A.G.; Sane, P.V.; Hurry, V.; Oquist, G.; Huner, N.P. Photosystem II reaction centre quenching: Mechanisms and physiological role. Photosynth. Res 2008, 98, 565–574. [Google Scholar]
- Nellaepalli, S.; Kodru, S.; Subramanyam, R. Effect of cold temperature on regulation of state transitions in Arabidopsis thaliana. J. Photochem. Photobiol. B 2012, 112, 23–30. [Google Scholar]
| Gene | Transgenic host | Source plant | Phenotype and effects | References |
|---|---|---|---|---|
| AtICE1 | Arabidopsis thaliana | Arabidopsis thaliana | Freezing tolerance; activation of CBF3/DREB1A | [21] |
| AtICE2 | Arabidopsis thaliana | Arabidopsis thaliana | Freezing tolerance; activation of CBF1/DREB1B | [23] |
| AtICE1 | Cucumis sativus | Arabidopsis thaliana | Chilling tolerance; dwarf | [32] |
| SlICE1 | Solanum lycopersicum | Solanum lycopersicum | Chilling tolerance; accumulation of antioxidants | [27,28] |
| TaICE141, TaICE187 | Arabidopsis thaliana | Triticum aestivum | Freezing tolerance | [29] |
| AtCBF1, AtCBF2, AtCBF3 | Arabidopsis thaliana | Arabidopsis thaliana | Freezing, salt and drought tolerance; constitutive expression of COR | [20,33,34] |
| OsDREB1A, OsDREB1B, OsDREB1C | Oryza sativa | Oryza sativa | Chilling, salt and drought tolerance; dwarf | [35] |
| HvCBF4 | Oryza sativa | Hordeum vulgare | Chilling, drought and salt tolerance | [36] |
| TaDREB2, TaDREB3 | Triticum aestivum | Triticum aestivum | Freezing and drought tolerance; dwarf | [37] |
| AtCBF1, AtCBF2, AtCBF3 | Brassica napus | Arabidopsis thaliana | Freezing tolerance; constitutive expression of COR | [38] |
| AtCBF1 | Fragaria ananassa | Arabidopsis thaliana | Freezing tolerance | [39] |
| AtCBF3 | Solanum tuberosum | Arabidopsis thaliana | Freezing tolerance | [40] |
| AtCBF1 | Populus tremula x alba | Arabidopsis thaliana | Freezing tolerance | [41] |
| AtCBF3 | Triticum aestivum | Arabidopsis thaliana | Freezing tolerance | [42] |
| AtCBF3 | Nicotiana tabacum | Arabidopsis thaliana | Freezing tolerance | [43] |
| SlCBF1 | Arabidopsis thaliana | Solanum lycopersicum | Freezing tolerance | [44] |
| OsDREB1A | Arabidopsis thaliana | Oryza sativa | Freezing, drought and salt tolerance | [45] |
| ZmDREB1A | Arabidopsis thaliana | Zea mays | Freezing and drought tolerance; dwarf | [46] |
| VrCBF1, VrCBF4 | Arabidopsis thaliana | Vitis riparia | Freezing and drought tolerance; dwarf | [47] |
| HvCBF3 | Arabidopsis thaliana | Hordeum vulgare | Freezing tolerance | [48] |
| LpCBF3 | Arabidopsis thaliana | Lolium perenne | Freezing tolerance; dwarf | [49,50] |
| SlCBF1 | Arabidopsis thaliana | Solanum lycopersicum | Chilling and oxidative tolerance | [51] |
| MbDREB1 | Arabidopsis thaliana | Malus baccata | Chilling, drought and salt tolerance | [52] |
| GmDREB3 | Arabidopsis thaliana | Glycine max | Freezing, drought and salt tolerance | [53] |
| BpCBF1 | Arabidopsis thaliana | Betula pendula | Freezing tolerance; dwarf | [54] |
| OsDREB1B | Nicotiana plumbaginifolia | Oryza sativa | Freezing, oxidative and drought tolerance; disease resistance | [55] |
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Miura, K.; Furumoto, T. Cold Signaling and Cold Response in Plants. Int. J. Mol. Sci. 2013, 14, 5312-5337. https://doi.org/10.3390/ijms14035312
Miura K, Furumoto T. Cold Signaling and Cold Response in Plants. International Journal of Molecular Sciences. 2013; 14(3):5312-5337. https://doi.org/10.3390/ijms14035312
Chicago/Turabian StyleMiura, Kenji, and Tsuyoshi Furumoto. 2013. "Cold Signaling and Cold Response in Plants" International Journal of Molecular Sciences 14, no. 3: 5312-5337. https://doi.org/10.3390/ijms14035312




