Expression, Purification and Identification of CtCVNH, a Novel Anti-HIV (Human Immunodeficiency Virus) Protein from Ceratopteris thalictroides
Abstract
:1. Introduction
2. Results
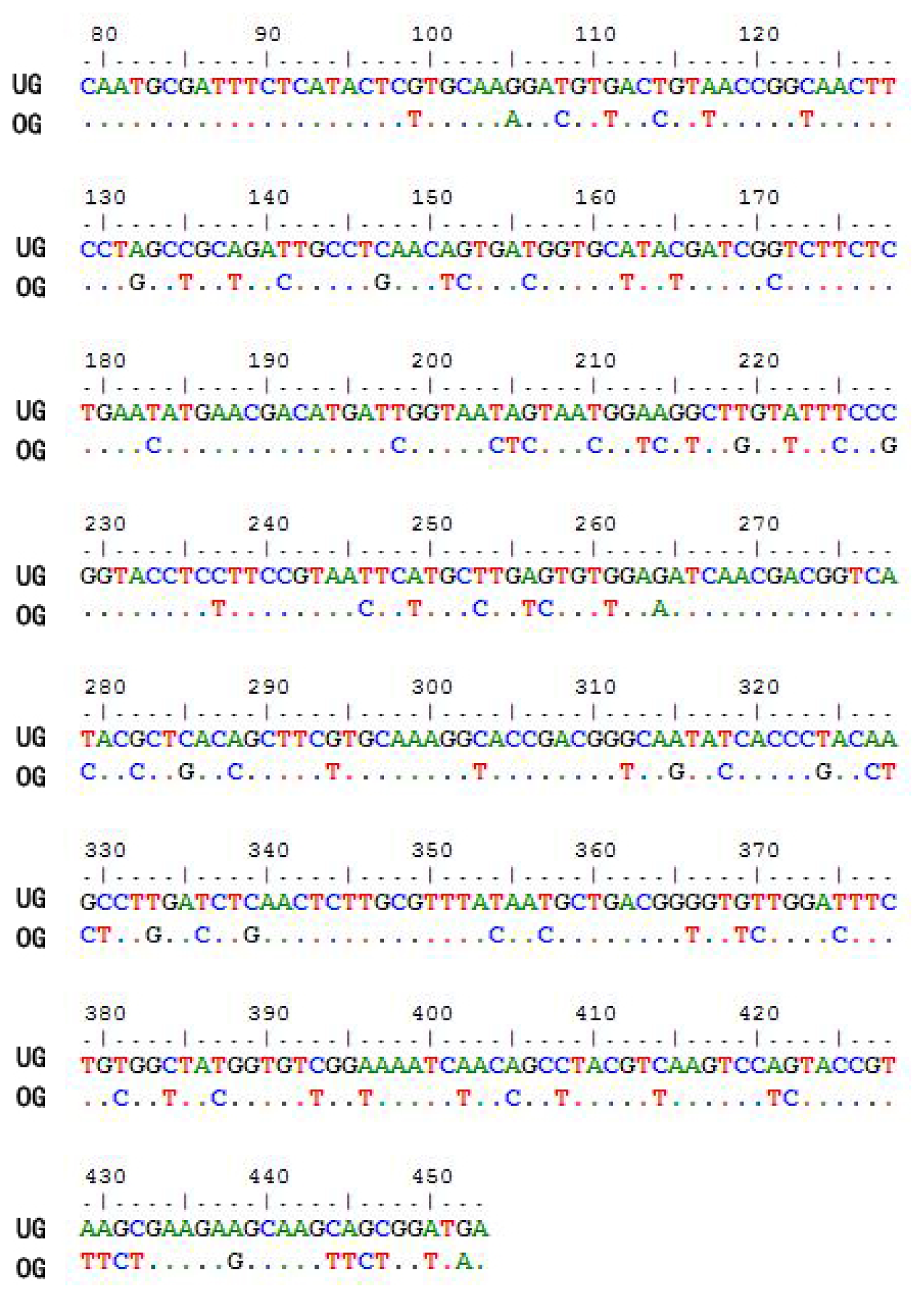
2.1. Optimization of the CtCVNH Gene
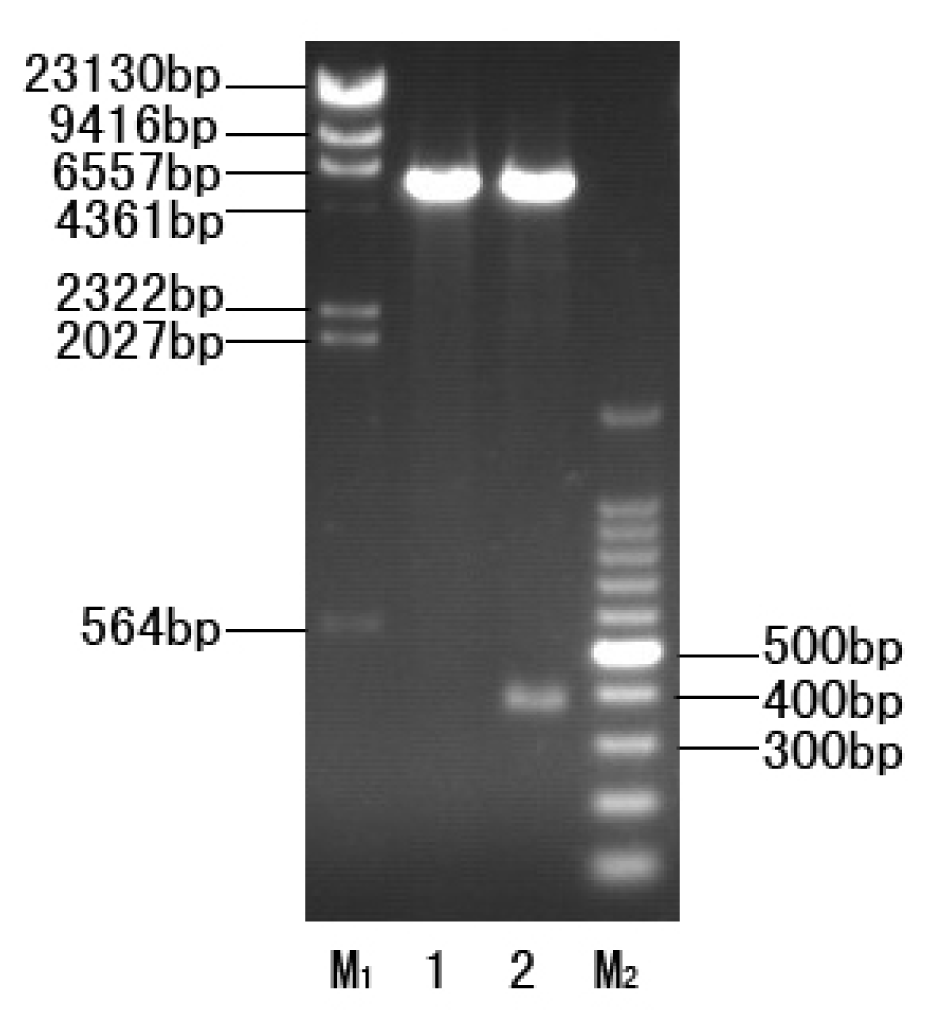
2.2. Construction of the Expression Strain
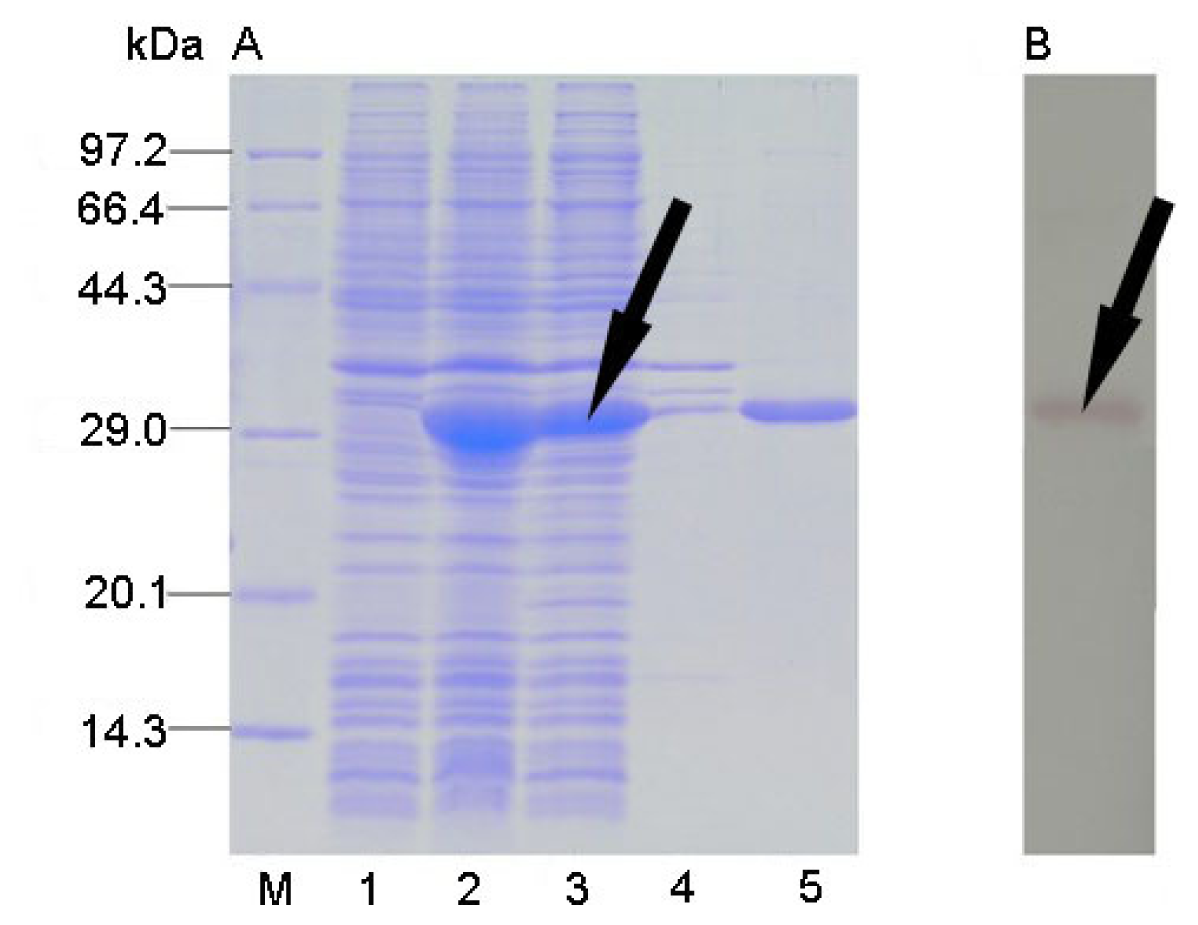
2.3. Protein Expression and Solubility Analysis
2.4. Protein Purification and Western Blot Analysis
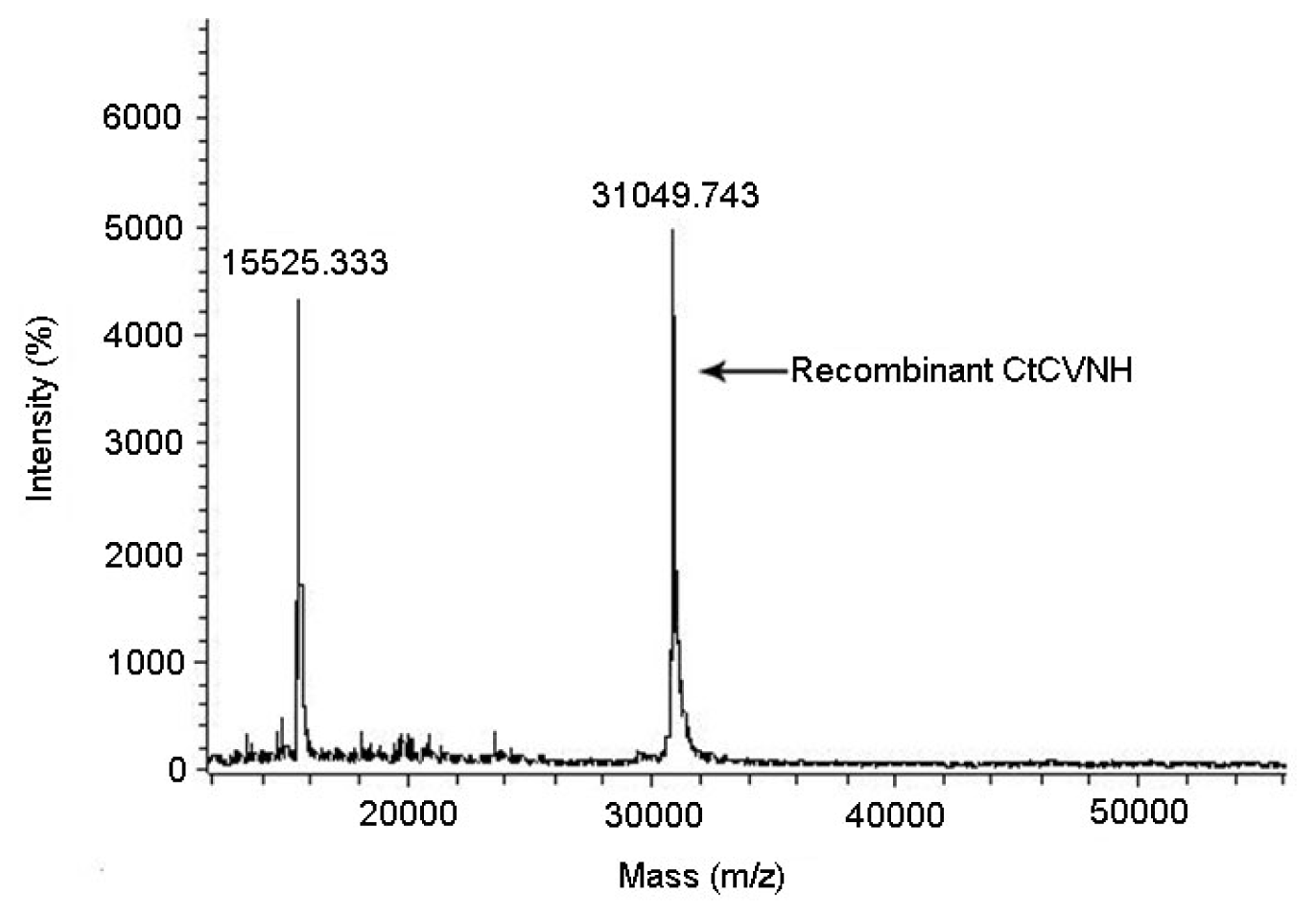
2.5. PMF Analysis and Molecular Weight Determination
3. Discussion
4. Experimental Section
4.1. Experimental Materials
4.2. Sequence Optimization and Construct Design
4.3. Construction of the Expression Strain
4.4. Expression of Recombinant Protein and Solubility Analysis
4.5. Protein Purification
4.6. Western Blot Analysis
4.7. Peptide Mass Fingerprinting Analysis and Molecular Weight Determination
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Clercq, E.D. Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int. J. Antimicrob. Agents 2009, 33, 307–320. [Google Scholar]
- Arts, E.J.; Hazuda, D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med 2012, 2, a007161. [Google Scholar]
- Boyd, M.R.; Gustafson, K.R.; Mcmahon, J.B.; Shoemaker, R.H.; O’Keefe, B.R.; Mori, T.; Gulakowski, R.J.; Wu, L.; Rivera, M.I.; Laurencot, C.M.; et al. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: Potential applications to microbicide development. Antimicrob. Agents Chemother 1997, 41, 1521–1530. [Google Scholar]
- Gustafson, K.R.; Sowder, R.C.; Henderson, L.E.; Cardellina, J.H.; McMahon, J.B.; Rajamani, U.; Pannell, L.K.; Boyd, M.R. Isolation, primary sequence determination, and disulfide bond structure of cyanovirin-N, an anti-HIV (human immunodeficiency virus) protein from the cyanobacterium Nostoc ellipsosporum. Biochem. Biophs. Res. Commun 1997, 238, 223–228. [Google Scholar]
- Yang, F.; Bewley, C.A.; Louis, J.M.; Gustafson, K.R.; Boyd, M.R.; Gronenborn, A.M.; Clore, G.M.; Wlodawer, A. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J. Mol. Biol 1999, 288, 403–412. [Google Scholar]
- Bewley, C.A.; Otero-Quintero, S. The potent anti-HIV protein cyanovirin-N contains two novel carbohydrate binding sites that selectively bind to Man8 D1D3 and Man9 with nanomolar affinity: Implications for binding to the HIV envelope protein gp120. J. Am. Chem. Soc 2001, 123, 3892–3902. [Google Scholar]
- Bewley, C.A.; Gustafson, K.R.; Boyd, M.R.; Covell, D.G.; Bax, A.; Clore, G.M.; Gronenborn, A.M. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat. Struct. Biol 1998, 5, 571–578. [Google Scholar]
- Bolmstedet, A.J.; O’Keefe, B.R.; Shenoy, S.R.; McMahon, J.B.; Boyd, M.R. Cyanovirin-N defines a new class of antiviral agent targeting N-linked, high-mannose glycans in an oligosaccharide-specific manner. Mol. Pharmacol 2005, 59, 949–954. [Google Scholar]
- Barrientos, L.G.; O’Keefe, B.R.; Bray, M.; Sanchez, A.; Gronenborn, A.M.; Boyd, M.R. Cyanovirin-N binds to the viral surface glycoprotein, GP1,2 and inhibits infectivity of Ebola virus. Antiviral. Res 2003, 58, 47–56. [Google Scholar]
- O’Keefe, B.R.; Smee, D.F.; Turpin, J.A.; Saucedo, C.J.; Gustafson, K.R.; Mori, T.; Blakeslee, D.; Buckheit, R.; Boyd, M.R. Potent anti-influenza activity of cyanovirin-N and interactions with viral hemagglutinin. Antimicrob. Agents Chemother 2003, 47, 2518–2525. [Google Scholar]
- Percudani, R.; Montanini, B.; Ottonello, S. The anti-HIV cyanovirin-N domain is evolutionarily conserved and occurs as a protein module in eukaryotes. Proteins 2005, 60, 670–6478. [Google Scholar]
- Qi, X.Q.; Yang, Y.X.; Su, Y.J.; Wang, T. Molecular cloning and sequence analysis of cyanovirin-N homology gene in Ceratopteris thalictroides. Am. Fern J 2009, 99, 78–92. [Google Scholar]
- Baneyx, F. Recombinant protein expression in Escherichia coli. Curr. Opin. Biotechnol 1999, 10, 411–421. [Google Scholar]
- Wong, S.H. Cloning of flavin reductase into pET32a (+) expression vector lacking the thioredoxin A tag to study solubility of EDTA monooxygenase A in overexpression systems. J. Exp. Microbiol. Immunol 2005, 8, 59–66. [Google Scholar]
- Burgess-Brown, N.A.; Sharma, S.; Sobot, F.; Loenarz, C.; Oppermann, U.; Gileadi, O. Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expr. Purif 2008, 59, 94–102. [Google Scholar]
- Han, J.H.; Choi, Y.S.; Kim, W.J.; Jeon, Y.H.; Lee, S.K.; Lee, B.J.; Ryu, K.S. Codon optimization enhances protein expression of human peptide deformylase in E. coli. Protein Expr. Purif 2010, 70, 224–230. [Google Scholar]
- Stewart, E.J.; Aslund, F.; Beckwith, J. Disulfide bond formation in the Escherichia coli cytoplasm: An in vivo role reversal for the thioredoxins. EMBO J 1998, 17, 5543–5550. [Google Scholar]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar]
- Grote, A.; Hiller, K.; Scheer, M.; Munch, R.; Nortemann, B.; Hempet, D.C.; Jahn, D. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 2005, 33, 526–531. [Google Scholar]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res 2008, 36, 70–74. [Google Scholar]
- Li, N.; Sun, J.B.; W. J.; Q. L. G.; W. T.; Su, Y.J. Optimization for culture condition of CVNH in Escherichia coli, A novel anti-HIV (Human Immunodeficiency Virus) protein (in Chinese). Acta Sci. Nat. Univ. Sunyatseni 2013, 52, 90–96. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Sun, J.; Su, Y.; Wang, T. Expression, Purification and Identification of CtCVNH, a Novel Anti-HIV (Human Immunodeficiency Virus) Protein from Ceratopteris thalictroides. Int. J. Mol. Sci. 2013, 14, 7506-7514. https://doi.org/10.3390/ijms14047506
Sun J, Su Y, Wang T. Expression, Purification and Identification of CtCVNH, a Novel Anti-HIV (Human Immunodeficiency Virus) Protein from Ceratopteris thalictroides. International Journal of Molecular Sciences. 2013; 14(4):7506-7514. https://doi.org/10.3390/ijms14047506
Chicago/Turabian StyleSun, Junbo, Yingjuan Su, and Ting Wang. 2013. "Expression, Purification and Identification of CtCVNH, a Novel Anti-HIV (Human Immunodeficiency Virus) Protein from Ceratopteris thalictroides" International Journal of Molecular Sciences 14, no. 4: 7506-7514. https://doi.org/10.3390/ijms14047506



