Tailoring the Models of Transcription
Abstract
:1. Introduction
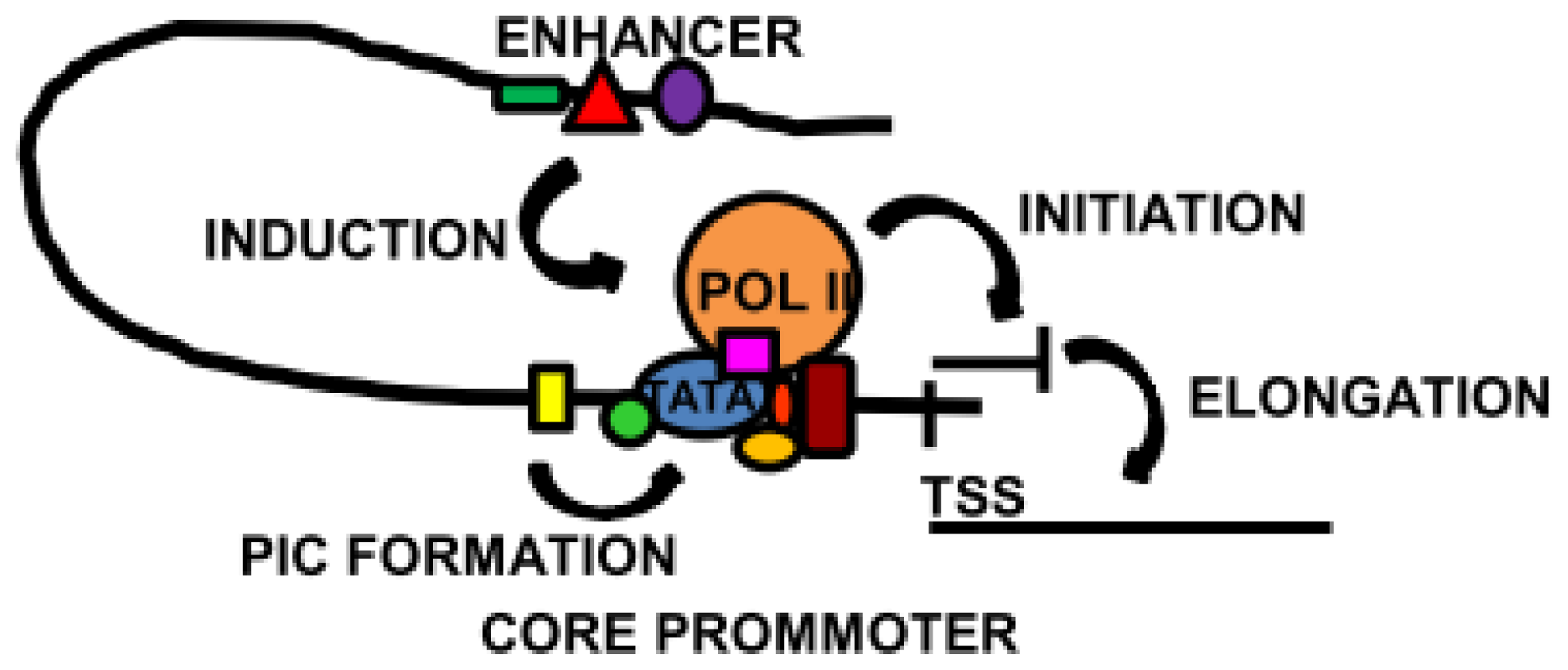
2. Regulatory Sequences: Promoters
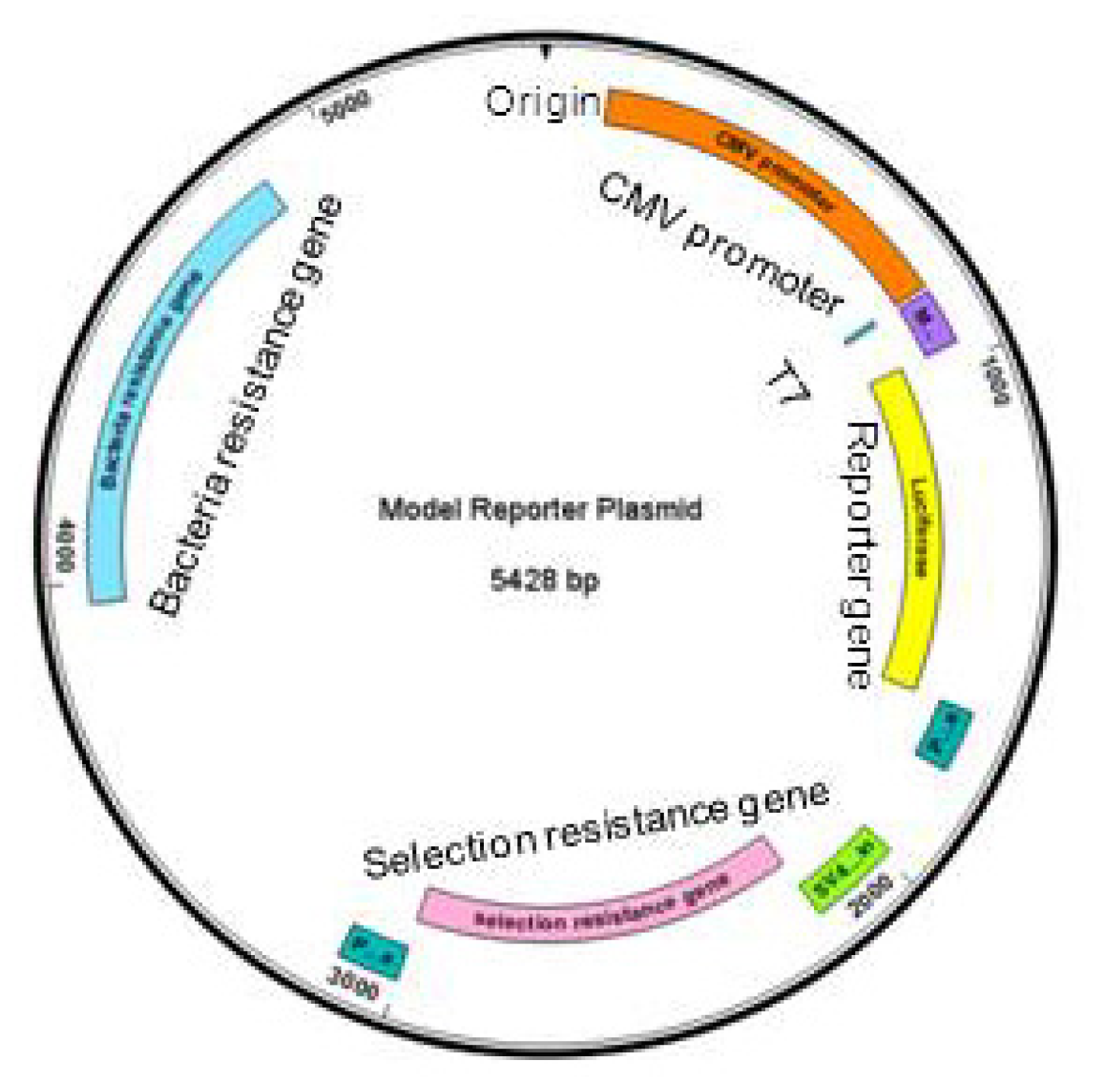
2.1. Pasting Reporter Genes
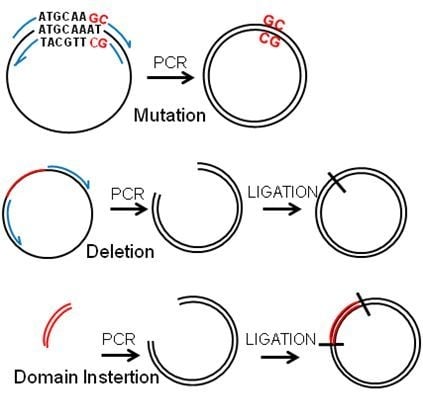
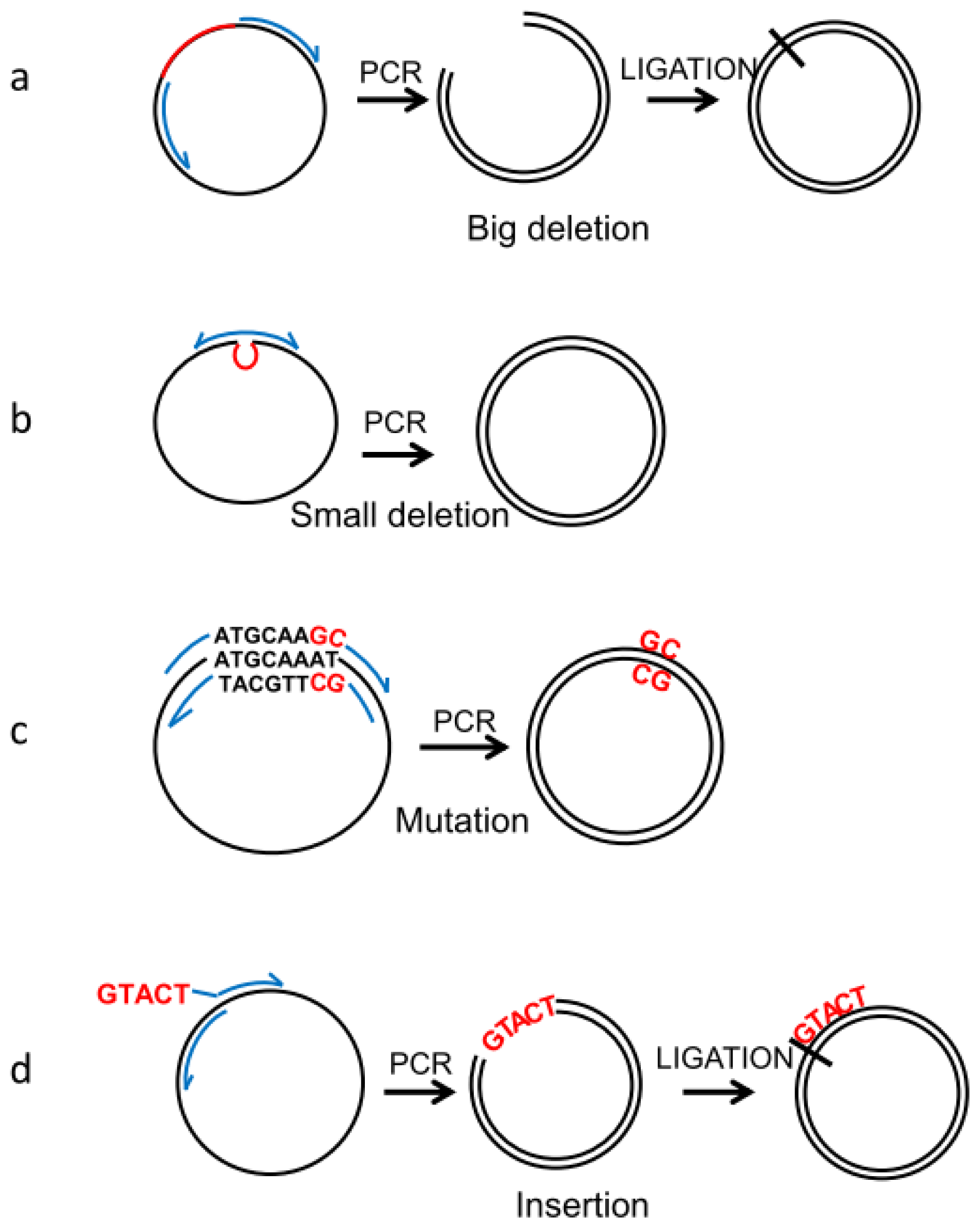
2.2. Deletions and Mutations
2.3. Insertions
3. Regulatory Genes: Transcription Factors
3.1. Modifications
3.2. Dominant Negative
3.3. Pasting Domains
4. Live Reporters
5. Concluding Remarks
Acknowledgements
References
- Nechaev, S.; Adelman, K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 2011, 1809, 34–45. [Google Scholar]
- Margaritis, T.; Holstege, F.C. Poised RNA polymerase II gives pause for thought. Cell 2008, 133, 581–584. [Google Scholar]
- Core, L.J.; Waterfall, J.J.; Lis, J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 2008, 322, 1845–1848. [Google Scholar]
- Sng, J.C.; Taniura, H.; Yoneda, Y. A tale of early response genes. Biol. Pharm. Bull 2004, 27, 606–612. [Google Scholar]
- Hargreaves, D.C.; Horng, T.; Medzhitov, R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 2009, 138, 129–145. [Google Scholar]
- Van Guilder, H.D.; Vrana, K.E.; Freeman, W.M. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 2008, 44, 619–626. [Google Scholar]
- Slonim, D.K.; Yanai, I. Getting started in gene expression microarray analysis. PLoS Comput. Biol 2009, 5, e1000543. [Google Scholar]
- Bertone, P.; Stolc, V.; Royce, T.E.; Rozowsky, J.S.; Urban, A.E.; Zhu, X.; Rinn, J.L.; Tongprasit, W.; Samanta, M.; Weissman, S.; et al. Global identification of human transcribed sequences with genome tiling arrays. Science 2004, 306, 2242–2246. [Google Scholar]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev 2011, 12, 87–98. [Google Scholar]
- Alam, J.; Cook, J.L. Reporter genes: Application to the study of mammalian gene transcription. Anal. Biochem 1990, 188, 245–254. [Google Scholar]
- Colbert, R.A.; Balbi, D.; Johnson, A.; Bailey, J.A.; Allen, J.M. Vasoactive intestinal peptide stimulates neuropeptide Y gene expression and causes neurite extension in PC12 cells through independent mechanisms. J. Neurosci 1994, 14, 7141–7147. [Google Scholar]
- Balbi, D.; Allen, J.M. Role of protein kinase C in mediating NGF effect on neuropeptide Y expression in PC12 cells. Brain Res. Mol. Brain Res 1994, 23, 310–316. [Google Scholar]
- Walker, M.D.; Edlund, T.; Boulet, A.M.; Rutter, W.J. Cell-specific expression controlled by the 5′-flanking region of insulin and chymotrypsin genes. Nature 1983, 306, 557–561. [Google Scholar]
- Balu, B.; Blair, P.L.; Adams, J.H. Identification of the transcription initiation site reveals a novel transcript structure for Plasmodium falciparum maebl. Exp. Parasitol 2009, 121, 110–114. [Google Scholar]
- Williams, T.M.; Burlein, J.E.; Ogden, S.; Kricka, L.J.; Kant, J.A. Advantages of firefly luciferase as a reporter gene: Application to the interleukin-2 gene promoter. Anal. Biochem 1989, 176, 28–32. [Google Scholar]
- De Wet, J.R.; Wood, K.V.; DeLuca, M.; Helinski, D.R.; Subramani, S. Firefly luciferase gene: Structure and expression in mammalian cells. Mol. Cell Biol 1987, 7, 725–737. [Google Scholar]
- De Wet, J.R.; Wood, K.V.; Helinski, D.R.; DeLuca, M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc. Natl. Acad. Sci. USA 1985, 82, 7870–7873. [Google Scholar]
- Bentrari, F.; Arnould, L.; Jackson, A.P.; Jeannin, J.F.; Pance, A. Progesterone enhances cytokine-stimulated nitric oxide synthase II expression and cell death in human breast cancer cells. Lab. Investig. J. Tech. Methods Pathol 2005, 85, 624–632. [Google Scholar]
- Logette, E.; Wotawa, A.; Solier, S.; Desoche, L.; Solary, E.; Corcos, L. The human caspase-2 gene: Alternative promoters, pre-mRNA splicing and AUG usage direct isoform-specific expression. Oncogene 2003, 22, 935–946. [Google Scholar]
- Toh, W.H.; Logette, E.; Corcos, L.; Sabapathy, K. TAp73beta and DNp73beta activate the expression of the pro-survival caspase-2S. Nucleic Acids Res 2008, 36, 4498–4509. [Google Scholar]
- Wang, Q.E.; Han, C.; Zhang, B.; Sabapathy, K.; Wani, A.A. Nucleotide excision repair factor XPC enhances DNA damage-induced apoptosis by downregulating the antiapoptotic short isoform of caspase-2. Cancer Res 2012, 72, 666–675. [Google Scholar]
- de Vera, M.E.; Shapiro, R.A.; Nussler, A.K.; Mudgett, J.S.; Simmons, R.L.; Morris, S.M., Jr; Billiar, T.R.; Geller, D.A. Transcriptional regulation of human inducible nitric oxide synthase (NOS2) gene by cytokines: Initial analysis of the human NOS2 promoter. Proc. Natl. Acad. Sci. USA 1996, 93, 1054–1059. [Google Scholar]
- Taylor, B.S.; de Vera, M.E.; Ganster, R.W.; Wang, Q.; Shapiro, R.A.; Morris, S.M., Jr; Billiar, T.R.; Geller, D.A. Multiple NF-kappaB enhancer elements regulate cytokine induction of the human inducible nitric oxide synthase gene. J. Biol. Chem. 1998, 273, 15148–15156. [Google Scholar]
- Guo, Z.; Shao, L.; Du, Q.; Park, K.S.; Geller, D.A. Identification of a classic cytokine-induced enhancer upstream in the human iNOS promoter. FASEB J 2007, 21, 535–542. [Google Scholar]
- Pance, A.; Chantome, A.; Reveneau, S.; Bentrari, F.; Jeannin, J.F. A repressor in the proximal human inducible nitric oxide synthase promoter modulates transcriptional activation. FASEB J 2002, 16, 631–633. [Google Scholar]
- Gomez-Benito, M.; Loayza-Puch, F.; Oude Vrielink, J.A.; Odero, M.D.; Agami, R. 3′UTR-mediated gene silencing of the Mixed Lineage Leukemia (MLL) gene. PLoS One 2011, 6, e25449. [Google Scholar]
- Cai, Y.H.; Huang, H. Advances in the study of protein-DNA interaction. Amino Acids 2012, 43, 1141–1146. [Google Scholar]
- Kristof, A.S.; Marks-Konczalik, J.; Moss, J. Mitogen-activated protein kinases mediate activator protein-1-dependent human inducible nitric-oxide synthase promoter activation. J. Biol. Chem 2001, 276, 8445–8452. [Google Scholar]
- Park, K.S.; Guo, Z.; Shao, L.; Du, Q.; Geller, D.A. A far-upstream Oct-1 motif regulates cytokine-induced transcription of the human inducible nitric oxide synthase gene. J. Mol. Biol 2009, 390, 595–603. [Google Scholar]
- Reveneau, S.; Petrakis, T.G.; Goldring, C.E.; Chantome, A.; Jeannin, J.F.; Pance, A. Oct-1 cooperates with the TATA binding initiation complex to control rapid transcription of human iNOS. Cell Mol. Life Sci 2012, 69, 2609–2619. [Google Scholar]
- Pance, A.; Livesey, F.J.; Jackson, A.P. A role for the transcriptional repressor REST in maintaining the phenotype of neurosecretory-deficient PC12 cells. J. Neurochem 2006, 99, 1435–1444. [Google Scholar]
- Bournat, J.C.; Allen, J.M. Regulation of the Y1 neuropeptide Y receptor gene expression in PC12 cells. Brain Res. Mol. Brain Res 2001, 90, 149–164. [Google Scholar]
- Carter, A.B.; Knudtson, K.L.; Monick, M.M.; Hunninghake, G.W. The p38 mitogen-activated protein kinase is required for NF-kappaB-dependent gene expression. The role of TATA-binding protein (TBP). J. Biol. Chem 1999, 274, 30858–30863. [Google Scholar]
- Chantome, A.; Pance, A.; Gauthier, N.; Vandroux, D.; Chenu, J.; Solary, E.; Jeannin, J.F.; Reveneau, S. Casein kinase II-mediated phosphorylation of NF-kappaB p65 subunit enhances inducible nitric-oxide synthase gene transcription in vivo. J. Biol. Chem 2004, 279, 23953–23960. [Google Scholar]
- Schoenherr, C.J.; Paquette, A.J.; Anderson, D.J. Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. USA 1996, 93, 9881–9886. [Google Scholar]
- Bertolino, E.; Singh, H. POU/TBP cooperativity: A mechanism for enhancer action from a distance. Mol. Cell 2002, 10, 397–407. [Google Scholar]
- Marsman, J.; Horsfield, J.A. Long distance relationships: Enhancer-promoter communication and dynamic gene transcription. Biochim. Biophys. Acta 2012, 1819, 1217–1227. [Google Scholar]
- Dikstein, R. The unexpected traits associated with core promoter elements. Transcription 2011, 2, 201–206. [Google Scholar]
- Verrijzer, C.P.; van Oosterhout, J.A.; van Weperen, W.W.; van der Vliet, P.C. POU proteins bend DNA via the POU-specific domain. EMBO J 1991, 10, 3007–3014. [Google Scholar]
- Stadhouders, R.; Kolovos, P.; Brouwer, R.; Zuin, J.; van den Heuvel, A.; Kockx, C.; Palstra, R.J.; Wendt, K.S.; Grosveld, F.; van Ijcken, W.; et al. Multiplexed chromosome conformation capture sequencing for rapid genome-scale high-resolution detection of long-range chromatin interactions. Nat. Protocols 2013, 8, 509–524. [Google Scholar]
- Tolhuis, B.; Palstra, R.J.; Splinter, E.; Grosveld, F.; de Laat, W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 2002, 10, 1453–1465. [Google Scholar]
- Shah, P.C.; Bertolino, E.; Singh, H. Using altered specificity Oct-1 and Oct-2 mutants to analyze the regulation of immunoglobulin gene transcription. EMBO J 1997, 16, 7105–7117. [Google Scholar]
- Ooi, L.; Wood, I.C. Chromatin crosstalk in development and disease: Lessons from REST. Nat. Rev 2007, 8, 544–554. [Google Scholar]
- Wood, I.C.; Belyaev, N.D.; Bruce, A.W.; Jones, C.; Mistry, M.; Roopra, A.; Buckley, N.J. Interaction of the repressor element 1-silencing transcription factor (REST) with target genes. J. Mol. Biol 2003, 334, 863–874. [Google Scholar]
- Hao, H. Genome-wide occupancy analysis by ChIP-chip and ChIP-Seq. Adv. Exp. Med. Biol 2012, 723, 753–759. [Google Scholar]
- Bruce, A.W.; Donaldson, I.J.; Wood, I.C.; Yerbury, S.A.; Sadowski, M.I.; Chapman, M.; Gottgens, B.; Buckley, N.J. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. USA 2004, 101, 10458–10463. [Google Scholar]
- Bruce, A.W.; Lopez-Contreras, A.J.; Flicek, P.; Down, T.A.; Dhami, P.; Dillon, S.C.; Koch, C.M.; Langford, C.F.; Dunham, I.; Andrews, R.M.; et al. Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res 2009, 19, 994–1005. [Google Scholar]
- Sadowski, I.; Ma, J.; Triezenberg, S.; Ptashne, M. GAL4-VP16 is an unusually potent transcriptional activator. Nature 1988, 335, 563–564. [Google Scholar]
- Greenwald, I. Notch and the awesome power of genetics. Genetics 2012, 191, 655–669. [Google Scholar]
- Struhl, G.; Adachi, A. Nuclear access and action of notch in vivo. Cell 1998, 93, 649–660. [Google Scholar]
- Struhl, G.; Greenwald, I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 1999, 398, 522–525. [Google Scholar]
- Zhang, Y.W.; Wang, R.; Liu, Q.; Zhang, H.; Liao, F.F.; Xu, H. Presenilin/gamma-secretase-dependent processing of beta-amyloid precursor protein regulates EGF receptor expression. Proc. Natl. Acad. Sci. USA 2007, 104, 10613–10618. [Google Scholar]
- Sikorski, T.W.; Joo, Y.J.; Ficarro, S.B.; Askenazi, M.; Buratowski, S.; Marto, J.A. Proteomic analysis demonstrates activator- and chromatin-specific recruitment to promoters. J. Biol. Chem 2012, 287, 35397–35408. [Google Scholar]
- Watanabe, Y.; Zaffran, S.; Kuroiwa, A.; Higuchi, H.; Ogura, T.; Harvey, R.P.; Kelly, R.G.; Buckingham, M. Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2–5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc. Natl. Acad. Sci. USA 2012, 109, 18273–18280. [Google Scholar]
- Mourikis, P.; Gopalakrishnan, S.; Sambasivan, R.; Tajbakhsh, S. Cell-autonomous Notch activity maintains the temporal specification potential of skeletal muscle stem cells. Development 2012, 139, 4536–4548. [Google Scholar]
- Chew, S.K.; Rad, R.; Futreal, P.A.; Bradley, A.; Liu, P. Genetic screens using the piggyBac transposon. Methods 2011, 53, 366–371. [Google Scholar]
- Rad, R.; Rad, L.; Wang, W.; Cadinanos, J.; Vassiliou, G.; Rice, S.; Campos, L.S.; Yusa, K.; Banerjee, R.; Li, M.A.; et al. PiggyBac transposon mutagenesis: A tool for cancer gene discovery in mice. Science 2010, 330, 1104–1107. [Google Scholar]
- Balu, B. Moving “forward” in plasmodium genetics through a transposon-based approach. J. Trop. Med 2012, 2012, 829210. [Google Scholar]
- Balu, B.; Maher, S.P.; Pance, A.; Chauhan, C.; Naumov, A.V.; Andrews, R.M.; Ellis, P.D.; Khan, S.M.; Lin, J.W.; Janse, C.J.; et al. CCR4-associated factor 1 coordinates the expression of Plasmodium falciparum egress and invasion proteins. Eukaryot. Cell 2011, 10, 1257–1263. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Pance, A. Tailoring the Models of Transcription. Int. J. Mol. Sci. 2013, 14, 7583-7597. https://doi.org/10.3390/ijms14047583
Pance A. Tailoring the Models of Transcription. International Journal of Molecular Sciences. 2013; 14(4):7583-7597. https://doi.org/10.3390/ijms14047583
Chicago/Turabian StylePance, Alena. 2013. "Tailoring the Models of Transcription" International Journal of Molecular Sciences 14, no. 4: 7583-7597. https://doi.org/10.3390/ijms14047583