Expression Pattern of Small Nucleolar RNA Host Genes and Long Non-Coding RNA in X-rays-Treated Lymphoblastoid Cells
Abstract
:1. Introduction
2. Results and Discussion
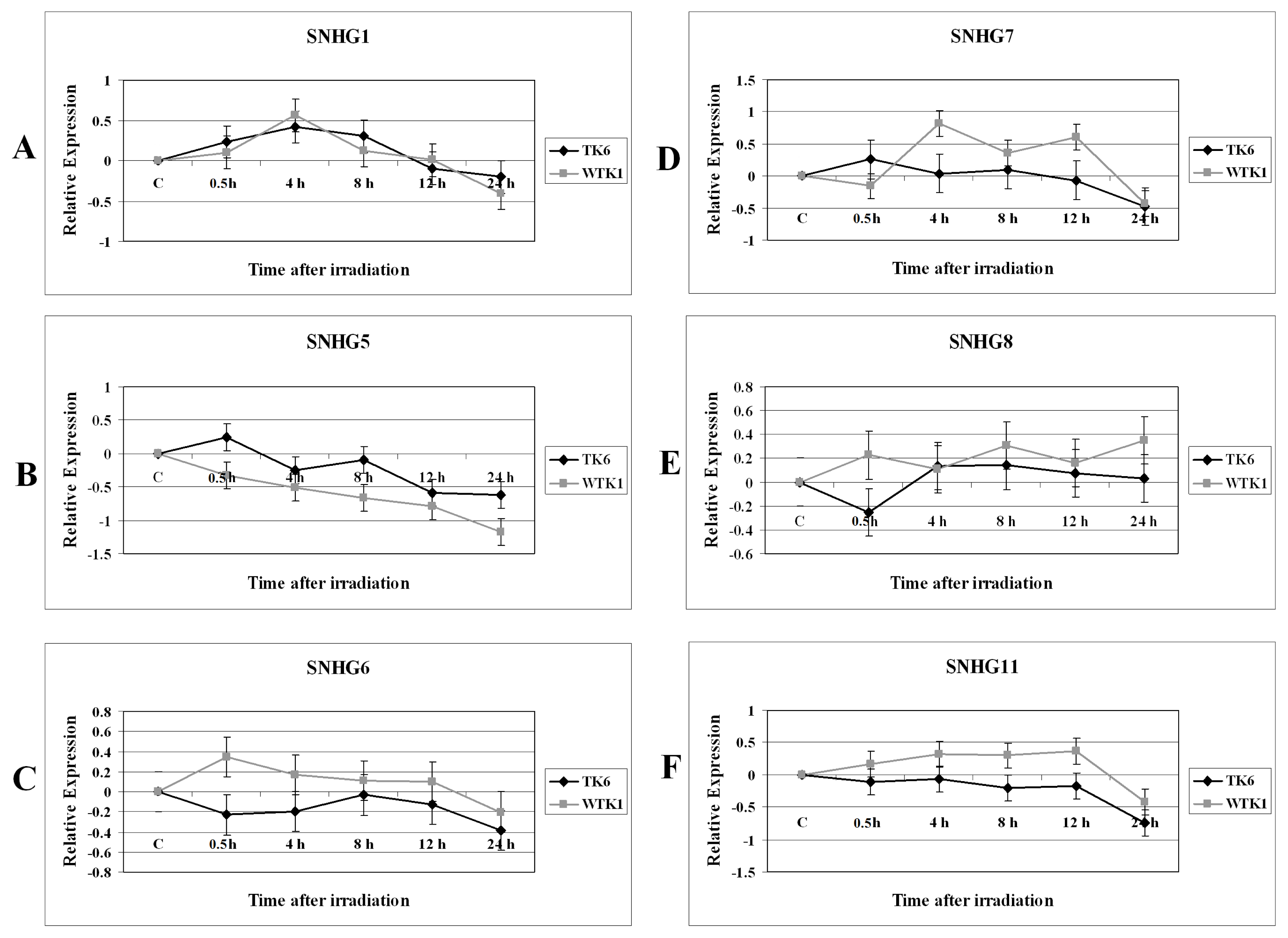
2.1. Results
2.2. Discussion
2.2.1. Expression Alterations of Small Nucleolar RNA Host Genes in X-rays-Treated Cells
2.2.2. Expression Changes of Long Non-Coding RNA in Irradiated Cells
3. Experimental Section
3.1. Cell Culture and Ionizing Radiation Treatment
3.2. Non-Coding RNA
3.3. cDNA Synthesis, Quantitative Real-Time Polymerase Chain Reaction (QPCR) and Data Analysis
4. Conclusions
Acknowledgements
References
- Yang, N.; Chaudhry, M.A.; Wallace, S.S. Base excision repair by hNTH1 and hOGG1: A two edged sword in the processing of DNA damage in gamma-irradiated human cells. DNA Repair (Amst) 2006, 5, 43–51. [Google Scholar]
- Chaudhry, M.A. Analysis of gene expression in normal and cancer cells exposed to gamma-radiation. J. Biomed. Biotechnol 2008, 2008, 541678. [Google Scholar]
- Chaudhry, M.A. Radiation-induced gene expression profile of human cells deficient in 8-hydroxy-2′-deoxyguanine glycosylase. Int. J. Cancer 2006, 118, 633–642. [Google Scholar]
- Chaudhry, M.A. Base excision repair of ionizing radiation-induced DNA damage in G1 and G2 cell cycle phases. Cancer Cell Int 2007, 7, 15. [Google Scholar]
- Chaudhry, M.A. Bystander effect: Biological endpoints and microarray analysis. Mutat. Res 2006, 597, 98–112. [Google Scholar]
- Chaudhry, M.A.; Omaruddin, R.A. Mitochondrial gene expression in directly irradiated and nonirradiated bystander cells. Cancer Biother. Radiopharm 2011, 26, 657–663. [Google Scholar]
- Chaudhry, M.A.; Omaruddin, R.A. Differential DNA methylation alterations in radiation-sensitive and -resistant cells. DNA Cell Biol 2012, 31, 908–916. [Google Scholar]
- Chaudhry, M.A.; Omaruddin, R.A. Transcriptional changes of mitochondrial genes in irradiated cells proficient or deficient in p53. J. Genet 2012, 91, 105–110. [Google Scholar]
- Chaudhry, M.A.; Omaruddin, R.A.; Kreger, B.; de Toledo, S.M.; Azzam, E.I. Micro RNA responses to chronic or acute exposures to low dose ionizing radiation. Mol. Biol. Rep 2012, 39, 7549–7558. [Google Scholar]
- Chaudhry, M.A. Real-time PCR analysis of micro-RNA expression in ionizing radiation-treated cells. Cancer Biother. Radiopharm 2009, 24, 49–56. [Google Scholar]
- Templin, T.; Paul, S.; Amundson, S.A.; Young, E.F.; Barker, C.A.; Wolden, S.L.; Smilenov, L.B. Radiation-induced micro-RNA expression changes in peripheral blood cells of radiotherapy patients. Int. J. Radiat. Oncol. Biol. Phys 2011, 80, 549–557. [Google Scholar]
- Chaudhry, M.A.; Omaruddin, R.A. Differential regulation of microRNA expression in irradiated and bystander cells. Mol. Biol. (Mosk) 2012, 46, 634–643. [Google Scholar]
- Lhakhang, T.W.; Chaudhry, M.A. Interactome of Radiation-Induced microRNA-Predicted Target Genes. Comp. Funct. Genomics 2012, 2012, 569731. [Google Scholar]
- Chaudhry, M.A.; Sachdeva, H.; Omaruddin, R.A. Radiation-induced micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol 2010, 29, 553–561. [Google Scholar]
- Chaudhry, M.A.; Kreger, B.; Omaruddin, R.A. Transcriptional modulation of micro-RNA in human cells differing in radiation sensitivity. Int. J. Radiat. Biol 2010, 86, 569–583. [Google Scholar]
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar]
- Chen, L.L.; Carmichael, G.G. Long noncoding RNAs in mammalian cells: What, where, and why? WIREs RNA 2010, 1, 2–21. [Google Scholar]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev 2009, 23, 1494–1504. [Google Scholar]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet 2009, 10, 155–159. [Google Scholar]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol 2011, 12, 246–258. [Google Scholar]
- Siddiqi, S.; Matushansky, I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J. Cell Biochem 2012, 113, 373–380. [Google Scholar]
- Harvey, M.; Sands, A.T.; Weiss, R.S.; Hegi, M.E.; Wiseman, R.W.; Pantazis, P.; Giovanella, B.C.; Tainsky, M.A.; Bradley, A.; Donehower, L.A. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene 1993, 8, 2457–2467. [Google Scholar]
- Amundson, S.A.; Xia, F.; Wolfson, K.; Liber, H.L. Different cytotoxic and mutagenic responses induced by X-rays in two human lymphoblastoid cell lines derived from a single donor. Mutat. Res 1993, 286, 233–241. [Google Scholar]
- Xia, F.; Amundson, S.A.; Nickoloff, J.A.; Liber, H.L. Different capacities for recombination in closely related human lymphoblastoid cell lines with different mutational responses to X-irradiation. Mol. Cell Biol 1994, 14, 5850–5857. [Google Scholar]
- Tsai, M.H.; Chen, X.; Chandramouli, G.V.; Chen, Y.; Yan, H.; Zhao, S.; Keng, P.; Liber, H.L.; Coleman, C.N.; Mitchell, J.B.; et al. Transcriptional responses to ionizing radiation reveal that p53R2 protects against radiation-induced mutagenesis in human lymphoblastoid cells. Oncogene 2006, 25, 622–632. [Google Scholar]
- Xia, F.; Wang, X.; Wang, Y.-H.; Tsang, N.-M.; Yandell, D.W.; Kelsey, K.T.; Liber, H.L. Altered p53 Status Correlates with Differences in Sensitivity to Radiation-induced Mutation and Apoptosis in Two Closely Related Lymphoblast Lines. Cancer Res 1994, 55, 12–15. [Google Scholar]
- Meador, J.A.; Ghandhi, S.A.; Amundson, S.A. p53-independent downregulation of histone gene expression in human cell lines by high- and low-let radiation. Radiat. Res 2011, 175, 689–699. [Google Scholar]
- Bratkovic, T.; Rogelj, B. Biology and applications of small nucleolar RNAs. Cell Mol. Life Sci 2011, 68, 3843–3851. [Google Scholar]
- Dong, X.Y.; Guo, P.; Boyd, J.; Sun, X.; Li, Q.; Zhou, W.; Dong, J.T. Implication of snoRNA U50 in human breast cancer. J. Genet. Genomics 2009, 36, 447–454. [Google Scholar]
- Higa-Nakamine, S.; Suzuki, T.; Uechi, T.; Chakraborty, A.; Nakajima, Y.; Nakamura, M.; Hirano, N.; Suzuki, T.; Kenmochi, N. Loss of ribosomal RNA modification causes developmental defects in zebrafish. Nucleic Acids Res 2012, 40, 391–398. [Google Scholar]
- Holley, C.L.; Topkara, V.K. An introduction to small non-coding RNAs: miRNA and snoRNA. Cardiovasc. Drugs Ther 2011, 25, 151–159. [Google Scholar]
- Chaudhry, M.A.; Chodosh, L.A.; McKenna, W.G.; Muschel, R.J. Gene expression profile of human cells irradiated in G1 and G2 phases of cell cycle. Cancer Lett 2003, 195, 221–233. [Google Scholar]
- Valleron, W.; Laprevotte, E.; Gautier, E.F.; Quelen, C.; Demur, C.; Delabesse, E.; Agirre, X.; Prosper, F.; Kiss, T.; Brousset, P. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia 2012, 26, 2052–2060. [Google Scholar]
- Mei, Y.P.; Liao, J.P.; Shen, J.; Yu, L.; Liu, B.L.; Liu, L.; Li, R.Y.; Ji, L.; Dorsey, S.G.; Jiang, Z.R.; et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 2012, 31, 2794–2804. [Google Scholar]
- Huang, Y.; Liu, N.; Wang, J.P.; Wang, Y.Q.; Yu, X.L.; Wang, Z.B.; Cheng, X.C.; Zou, Q. Regulatory long non-coding RNA and its functions. J. Physiol. Biochem 2012, 68, 611–618. [Google Scholar]
- Niland, C.N.; Merry, C.R.; Khalil, A.M. Emerging Roles for Long Non-Coding RNAs in Cancer and Neurological Disorders. Front Genet 2012, 3, 25. [Google Scholar]
- Silva, J.M.; Perez, D.S.; Pritchett, J.R.; Halling, M.L.; Tang, H.; Smith, D.I. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics 2010, 95, 355–362. [Google Scholar]
- Mizutani, R.; Wakamatsu, A.; Tanaka, N.; Yoshida, H.; Tochigi, N.; Suzuki, Y.; Oonishi, T.; Tani, H.; Tano, K.; Ijiri, K.; et al. Identification and characterization of novel genotoxic stress-inducible nuclear long noncoding RNAs in mammalian cells. PLoS One 2012, 7, e34949. [Google Scholar]
- Maccani, M.A.; Knopik, V.S. Cigarette smoke exposure-associated alterations to non-coding RNA. Front Genet 2012, 3, 53. [Google Scholar]
- Schmidt, L.H.; Spieker, T.; Koschmieder, S.; Humberg, J.; Jungen, D.; Bulk, E.; Hascher, A.; Wittmer, D.; Marra, A.; Hillejan, L.; et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J. Thorac. Oncol 2011, 6, 1984–1992. [Google Scholar]
- Ozgur, E.; Mert, U.; Isin, M.; Okutan, M.; Dalay, N.; Gezer, U. Differential expression of long non-coding RNAs during genotoxic stress-induced apoptosis in HeLa and MCF-7 cells. Clin. Exp. Med. 2012. [Google Scholar] [CrossRef]
- Nakagawa, S.; Ip, J.Y.; Shioi, G.; Tripathi, V.; Zong, X.; Hirose, T.; Prasanth, K.V. Malat1 is not an essential component of nuclear speckles in mice. RNA 2012, 18, 1487–1499. [Google Scholar]
- Cooper, C.; Guo, J.; Yan, Y.; Chooniedass-Kothari, S.; Hube, F.; Hamedani, M.K.; Murphy, L.C.; Myal, Y.; Leygue, E. Increasing the relative expression of endogenous non-coding Steroid Receptor RNA Activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res 2009, 37, 4518–4531. [Google Scholar]
- Foulds, C.E.; Tsimelzon, A.; Long, W.; Le, A.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. Research resource: Expression profiling reveals unexpected targets and functions of the human steroid receptor RNA activator (SRA) gene. Mol. Endocrinol 2010, 24, 1090–1105. [Google Scholar]
- Amaral, P.P.; Neyt, C.; Wilkins, S.J.; Askarian-Amiri, M.E.; Sunkin, S.M.; Perkins, A.C.; Mattick, J.S. Complex architecture and regulated expression of the Sox2ot locus during vertebrate development. RNA 2009, 15, 2013–2027. [Google Scholar]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet 2006, 51, 1087–1099. [Google Scholar]
- Lu, Y.; Zhang, K.; Li, C.; Yao, Y.; Tao, D.; Liu, Y.; Zhang, S.; Ma, Y. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS One 2012, 7, e30999. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar]

| Assay ID | Entrez Gene ID | Gene Symbol | Gene Name | Transcribed snoRNA |
|---|---|---|---|---|
| Hs00411543_m1 | 23642 | SNHG1 | Small nucleolar RNA host gene 1 | U22, U27, U28, U29, U30, and U31 box C/D |
| Hs00761246_s1 | 387066 | SNHG5 | Small nucleolar RNA host gene 5 | U50 and U50B box C/D |
| Hs00417251_m1 | 641638 | SNHG6 | Small nucleolar RNA host gene 6 | HBII-276 (U87) box C/D |
| Hs00288592_m1 | 84973 | SNHG7 | Small nucleolar RNA host gene 7 | ACA17 and ACA43 box H/ACA |
| Hs01399325_g1 | 100093630 | SNHG8 | Small nucleolar RNA host gene 8 | ACA24 box H/ACA |
| Hs00738242_m1 | 128439 | SNHG11 | Small nucleolar RNA host gene 11 | ACA39 and ACA60 box H/ACA |
| Hs00273907_s1 | 378938 | MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 | - |
| Hs00251579_m1 | 9782 | MATR3 | Matrin 3 | U19 box H/ACA |
| Hs00288796_m1 | 10011 | SRA1 | Steroid receptor RNA activator 1 | - |
| Hs00415716_m1 | 347689 | SOX2OT | SOX2 overlapping transcript | - |
| Hs00402814_m1 | 440823 | MIAT | Myocardial infarction associated transcript | - |
| Hs01041735_m1 | 9271 | PIWIL1 | P-element induced wimpy testis like 1 | - |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chaudhry, M.A. Expression Pattern of Small Nucleolar RNA Host Genes and Long Non-Coding RNA in X-rays-Treated Lymphoblastoid Cells. Int. J. Mol. Sci. 2013, 14, 9099-9110. https://doi.org/10.3390/ijms14059099
Chaudhry MA. Expression Pattern of Small Nucleolar RNA Host Genes and Long Non-Coding RNA in X-rays-Treated Lymphoblastoid Cells. International Journal of Molecular Sciences. 2013; 14(5):9099-9110. https://doi.org/10.3390/ijms14059099
Chicago/Turabian StyleChaudhry, M. Ahmad. 2013. "Expression Pattern of Small Nucleolar RNA Host Genes and Long Non-Coding RNA in X-rays-Treated Lymphoblastoid Cells" International Journal of Molecular Sciences 14, no. 5: 9099-9110. https://doi.org/10.3390/ijms14059099




