Current Status of Biomarkers for Prostate Cancer
Abstract
:1. Introduction
2. Prostatic Acid Phosphatase and Prostate Specific Antigen Tests
Refinement of the PSA Test
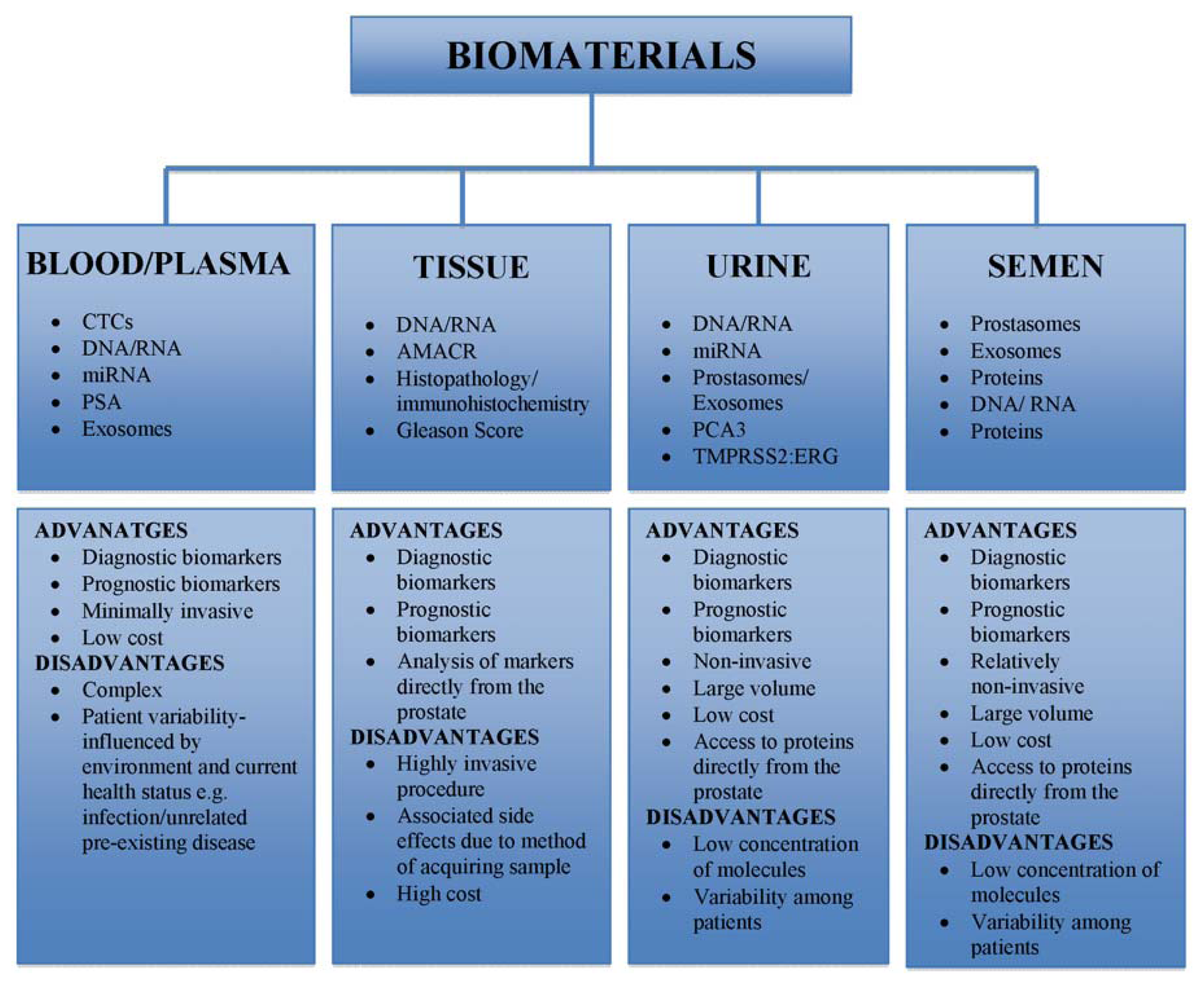
3. Biological Sampling and Tolerability
4. Current and Emerging Biomarkers
4.1. Genetic Markers of Prostate Cancer
4.2. Circulating miRNAs in Prostate Cancer
4.3. Protein-Based Biomarkers
4.4. Immunological Biomarkers
4.5. Microparticles
4.6. Circulating Tumor Cells
5. Conclusions
Conflict of Interest
References
- Thorne, H.; Willems, A.J.; Niedermayr, E.; Hoh, I.M.; Li, J.; Clouston, D.; Mitchell, G.; Fox, S.; Hopper, J.L.; Bolton, D. Decreased prostate cancer-specific survival of men with BRCA2 mutations from multiple breast cancer families. Cancer Prev. Res. (Phila) 2011, 4, 1002–1010. [Google Scholar]
- Prostate Cancer Foundation Australia. What is Prostate Cancer. Available online: http://www.prostate.org.au/articleLive/pages/What-is-Prostate-Cancer.html (accessed on 18 March 2013).
- Bangma, C.H.; Roemeling, S.; Schroder, F.H. Overdiagnosis and overtreatment of early detected prostate cancer. World J. Urol 2007, 25, 3–9. [Google Scholar]
- Assinder, S.J.; Nicholson, H. Prostate Disease: Prostate hyperplasia and prostate cancer and prostatitis. In Pathophysiology and Treatment of Male Sexual and Reproductive Dysfunction; Kandeel, E.F., Ed.; Marcel Dekker Inc: New York, NY, USA, 2007; pp. 423–439. [Google Scholar]
- Schroder, F.H.; Hugosson, J.; Roobol, M.J.; Tammela, T.L.; Ciatto, S.; Nelen, V.; Kwiatkowski, M.; Lujan, M.; Lilja, H.; Zappa, M.; et al. Screening and prostate-cancer mortality in a randomized European study. N. Engl. J. Med 2009, 360, 1320–1328. [Google Scholar]
- Moyer, V.A. Screening for prostate cancer: U.S. preventive services task force recommendation statement. Ann. Intern. Med 2012, 157, 120–134. [Google Scholar]
- Schroder, F.H. Stratifying risk—The U.S. preventive services task force and prostate-cancer screening. N. Engl. J. Med 2011, 365, 1953–1955. [Google Scholar]
- National Cancer Institute. NCI Dictionary of Cancer Terms. Available online: http://www.cancer.gov/dictionary?cdrid=45618 (accessed on 20 May 2013).
- Gutman, A.B.; Gutman, E.B. An “Acid” phosphatase occurring in the serum of patients with metastasizing carcinoma of the prostate gland. J. Clin. Invest 1938, 17, 473–478. [Google Scholar]
- Hernandez, J.; Thompson, I.M. Prostate-specific antigen: A review of the validation of the most commonly used cancer biomarker. Cancer 2004, 101, 894–904. [Google Scholar]
- Hara, M.; Koyanagi, Y.; Inoue, T.; Fukuyama, T. Some physico-chemical characteristics of “-seminoprotein”, an antigenic component specific for human seminal plasma. Forensic immunological study of body fluids and secretion. VII. NIhon Hogaku Zasshi 1971, 25, 322–324. [Google Scholar]
- Pinsky, P.F.; Kramer, B.S.; Crawford, E.D.; Grubb, R.L.; Urban, D.A.; Andriole, G.L.; Chia, D.; Levin, D.L.; Gohagan, J.K. Prostate volume and prostate-specific antigen levels in men enrolled in a large screening trial. Urology 2006, 68, 352–356. [Google Scholar]
- Liu, Y.; Hegde, P.; Zhang, F.; Hampton, G.; Jia, S. Prostate cancer—A biomarker perspective. Front. endocrinol 2012, 3, 72–79. [Google Scholar]
- Hessels, D.; Schalken, J.A. Urinary biomarkers for prostate cancer: A review. Asian J. Androl. 2013. [Google Scholar] [CrossRef]
- Cross, D.S.; Ritter, M.; Reding, D.J. Historical prostate cancer screening and treatment outcomes from a single institution. Clin. Med. Res 2012, 10, 97–105. [Google Scholar]
- Sardana, G.; Jung, K.; Stephan, C.; Diamandis, E.P. Proteomic analysis of conditioned media from the PC3, LNCaP, and 22Rv1 prostate cancer cell lines: Discovery and validation of candidate prostate cancer biomarkers. J. Proteome Res 2008, 7, 3329–3338. [Google Scholar]
- Mistry, K.; Cable, G. Meta-analysis of prostate-specific antigen and digital rectal examination as screening tests for prostate carcinoma. J. Am. Board Fam. Pract 2003, 16, 95–101. [Google Scholar]
- Schroder, F.H.; van der Cruijsen-Koeter, I.; de Koning, H.J.; Vis, A.N.; Hoedemaeker, R.F.; Kranse, R. Prostate cancer detection at low prostate specific antigen. J. Urol 2000, 163, 806–812. [Google Scholar]
- Lin, M.W.; Ho, J.W.; Harrison, L.C.; Dos Remedios, C.G.; Adelstein, S. An antibody-based leukocyte-capture microarray for the diagnosis of systemic lupus erythematosus. PLoS One 2013. [Google Scholar] [CrossRef]
- Haythorn, M.R.; Ablin, R.J. Prostate-specific antigen testing across the spectrum of prostate cancer. Biomark. Med 2011, 5, 515–526. [Google Scholar]
- Bodey, B.; Bodey, B., Jr; Kaiser, H.E. Immunocytochemical detection of prostate specific antigen expression in human primary and metastatic melanomas. Anticancer Res. 1997, 17, 2343–2346. [Google Scholar]
- Wilt, T.J.; Brawer, M.K.; Jones, K.M.; Barry, M.J.; Aronson, W.J.; Fox, S.; Gingrich, J.R.; Wei, J.T.; Gilhooly, P.; Grob, B.M.; et al. Radical prostatectomy versus observation for localized prostate cancer. N. Engl. J. Med 2012, 367, 203–213. [Google Scholar]
- Andriole, G.L.; Crawford, E.D.; Grubb, R.L., III; Buys, S.S.; Chia, D.; Church, T.R.; Fouad, M.N.; Isaacs, C.; Kvale, P.A.; Reding, D.J.; et al. Prostate cancer screening in the randomized prostate, lung, colorectal, and ovarian cancer screening trial: Mortality results after 13 years of follow-up. J. Natl. Cancer Inst. 2012, 104, 125–132. [Google Scholar]
- Roobol, M.J.; Zhu, X.; Schroder, F.H.; van Leenders, G.J.; van Schaik, R.H.; Bangma, C.H.; Steyerberg, E.W. A calculator for prostate cancer risk 4 years after an initially negative screen: Findings from ERSPC rotterdam. Eur. Urol 2012, 4, 627–633. [Google Scholar]
- Eckersberger, E.; Finkelstein, J.; Sadri, H.; Margreiter, M.; Taneja, S.S.; Lepor, H.; Djavan, B. Screening for prostate cancer: A review of the ERSPC and PLCO trials. Rev. Urol 2009, 11, 127–133. [Google Scholar]
- Gomella, L.G.; Liu, X.S.; Trabulsi, E.J.; Kelly, W.K.; Myers, R.; Showalter, T.; Dicker, A.; Wender, R. Screening for prostate cancer: The current evidence and guidelines controversy. Can. J. Urol 2011, 18, 5875–5883. [Google Scholar]
- Pierorazio, P.M.; Guzzo, T.J.; Han, M.; Bivalacqua, T.J.; Epstein, J.I.; Schaeffer, E.M.; Schoenberg, M.; Walsh, P.C.; Partin, A.W. Long-term survival after radical prostatectomy for men with high Gleason sum in pathologic specimen. Urology 2010, 76, 715–721. [Google Scholar]
- Resnick, M.J.; Koyama, T.; Fan, K.H.; Albertsen, P.C.; Goodman, M.; Hamilton, A.S.; Hoffman, R.M.; Potosky, A.L.; Stanford, J.L.; Stroup, A.M.; et al. Long-term functional outcomes after treatment for localized prostate cancer. N. Engl. J. Med 2013, 368, 436–445. [Google Scholar]
- Peyromaure, M.; Ravery, V.; Messas, A.; Toublanc, M.; Boccon-Gibod, L.; Boccon-Gibod, L. Pain and morbidity of an extensive prostate 10-biopsy protocol: A prospective study in 289 patients. J. Urol 2002, 167, 218–221. [Google Scholar]
- Seftel, A.D. Prostate cancer diagnosis is associated with an increased risk of erectile dysfunction after prostate biopsy. J. Urol 2012, 188, 2317–2318. [Google Scholar]
- Sardana, G.; Diamandis, E.P. Biomarkers for the diagnosis of new and recurrent prostate cancer. Biomark. Med 2012, 6, 587–596. [Google Scholar]
- Connolly, D.; Black, A.; Murray, L.J.; Napolitano, G.; Gavin, A.; Keane, P.F. Methods of calculating prostate-specific antigen velocity. Eur. Urol 2007, 52, 1044–1050. [Google Scholar]
- Shariat, S.F.; Semjonow, A.; Lilja, H.; Savage, C.; Vickers, A.J.; Bjartell, A. Tumor markers in prostate cancer I: Blood-based markers. Acta Oncol 2011, 50, 61–75. [Google Scholar]
- Zheng, X.Y.; Zhang, P.; Xie, L.P.; You, Q.H.; Cai, B.S.; Qin, J. Prostate-specific antigen velocity (PSAV) and PSAV per initial volume (PSAVD) for early detection of prostate cancer in Chinese men. Asian Pac. J. Cancer Prev 2012, 13, 5529–5533. [Google Scholar]
- Shariat, S.F.; Scardino, P.T.; Lilja, H. Screening for prostate cancer: An update. Can. J. Urol 2008, 15, 4363–4374. [Google Scholar]
- Auprich, M.; Augustin, H.; Budaus, L.; Kluth, L.; Mannweiler, S.; Shariat, S.F.; Fisch, M.; Graefen, M.; Pummer, K.; Chun, F.K. A comparative performance analysis of total prostate-specific antigen, percentage free prostate-specific antigen, prostate-specific antigen velocity and urinary prostate cancer gene 3 in the first, second and third repeat prostate biopsy. BJU Int 2012, 109, 1627–1635. [Google Scholar]
- Catalona, W.J.; Partin, A.W.; Slawin, K.M.; Brawer, M.K.; Flanigan, R.C.; Patel, A.; Richie, J.P.; deKernion, J.B.; Walsh, P.C.; Scardino, P.T.; et al. Use of the percentage of free prostate-specific antigen to enhance differentiation of prostate cancer from benign prostatic disease: A prospective multicenter clinical trial. J. Am. Med. Assoc 1998, 279, 1542–1547. [Google Scholar]
- Roddam, A.W.; Duffy, M.J.; Hamdy, F.C.; Ward, A.M.; Patnick, J.; Price, C.P.; Rimmer, J.; Sturgeon, C.; White, P.; Allen, N.E. Use of prostate-specific antigen (PSA) isoforms for the detection of prostate cancer in men with a PSA level of 2–10 ng/mL: Systematic review and meta-analysis. Eur. Urol 2005, 48, 386–399. [Google Scholar]
- Chun, F.K.; Suardi, N.; Capitanio, U.; Jeldres, C.; Ahyai, S.; Graefen, M.; Haese, A.; Steuber, T.; Erbersdobler, A.; Montorsi, F.; et al. Assessment of pathological prostate cancer characteristics in men with favorable biopsy features on predominantly sextant biopsy. Eur. Urol 2009, 55, 617–628. [Google Scholar]
- Capitanio, U.; Ahyai, S.; Graefen, M.; Jeldres, C.; Shariat, S.F.; Erbersdobler, A.; Schlomm, T.; Haese, A.; Steuber, T.; Heinzer, H.; et al. Assessment of biochemical recurrence rate in patients with pathologically confirmed insignificant prostate cancer. Urology 2008, 72, 1208–1211. [Google Scholar]
- Filella, X.; Gimenez, N. Evaluation of [−2]proPSA and Prostate Health Index (phi) for the detection of prostate cancer: A systematic review and meta-analysis. Clin. Chem. Lab. Med 2013, 51, 729–739. [Google Scholar]
- Nam, R.K.; Saskin, R.; Lee, Y.; Liu, Y.; Law, C.; Klotz, L.H.; Loblaw, D.A.; Trachtenberg, J.; Stanimirovic, A.; Simor, A.E.; et al. Increasing hospital admission rates for urological complications after transrectal ultrasound guided prostate biopsy. J. Urol 2013, 189, S12–S18. [Google Scholar]
- Loeb, S.; Carter, H.B.; Catalona, W.J.; Moul, J.W.; Schroder, F.H. Baseline prostate-specific antigen testing at a young age. Eur. Urol 2012, 61, 1–7. [Google Scholar]
- Nam, R.K.; Saskin, R.; Lee, Y.; Liu, Y.; Law, C.; Klotz, L.H.; Loblaw, D.A.; Trachtenberg, J.; Stanimirovic, A.; Simor, A.E.; et al. Increasing hospital admission rates for urological complications after transrectal ultrasound guided prostate biopsy. J. Urol 2010, 183, 963–968. [Google Scholar]
- Loeb, S.; van den Heuvel, S.; Zhu, X.; Bangma, C.H.; Schroder, F.H.; Roobol, M.J. Infectious complications and hospital admissions after prostate biopsy in a European randomized trial. Eur. Urol 2012, 61, 1110–1114. [Google Scholar]
- Loeb, S. Prostate biopsy: A risk-benefit analysis. J. Urol 2010, 183, 852–853. [Google Scholar]
- Goo, Y.A.; Goodlett, D.R. Advances in proteomic prostate cancer biomarker discovery. J. Proteomics 2010, 73, 1839–1850. [Google Scholar]
- Nedelkov, D. Mass spectrometry-based protein assays for in vitro diagnostic testing. Expert Rev. Mol. Diagn 2012, 12, 235–239. [Google Scholar]
- Wu, C.C.; Yates, J.R., III. The application of mass spectrometry to membrane proteomics. Nat. Biotechnol. 2003, 21, 262–267. [Google Scholar]
- Liu, Y.; Vlatkovic, L.; Saeter, T.; Servoll, E.; Waaler, G.; Nesland, J.M.; Giercksky, K.E.; Axcrona, K. Is the clinical malignant phenotype of prostate cancer a result of a highly proliferative immune-evasive B7-H3-expressing cell population? Int. J. Urol 2012, 19, 749–756. [Google Scholar]
- Mahnke, K.; Ring, S.; Johnson, T.S.; Schallenberg, S.; Schonfeld, K.; Storn, V.; Bedke, T.; Enk, A.H. Induction of immunosuppressive functions of dendritic cells in vivo by CD4+CD25+ regulatory T cells: Role of B7-H3 expression and antigen presentation. Eur. J. Immunol 2007, 37, 2117–2126. [Google Scholar]
- Roth, T.J.; Sheinin, Y.; Lohse, C.M.; Kuntz, S.M.; Frigola, X.; Inman, B.A.; Krambeck, A.E.; McKenney, M.E.; Karnes, R.J.; Blute, M.L.; et al. B7-H3 ligand expression by prostate cancer: A novel marker of prognosis and potential target for therapy. Cancer Res 2007, 67, 7893–7900. [Google Scholar]
- Zang, X.; Thompson, R.H.; Al-Ahmadie, H.A.; Serio, A.M.; Reuter, V.E.; Eastham, J.A.; Scardino, P.T.; Sharma, P.; Allison, J.P. B7-H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc. Natl. Acad. Sci. USA 2007, 104, 19458–19463. [Google Scholar]
- Gerdes, J.; Lemke, H.; Baisch, H.; Wacker, H.H.; Schwab, U.; Stein, H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol 1984, 133, 1710–1715. [Google Scholar]
- Madu, C.O.; Lu, Y. Novel diagnostic biomarkers for prostate cancer. J. Cancer 2010, 1, 150–177. [Google Scholar]
- Zhao, Z.; Zeng, G.; Zhong, W. Serum early prostate cancer antigen (EPCA) as a significant predictor of incidental prostate cancer in patients undergoing transurethral resection of the prostate for benign prostatic hyperplasia. Prostate 2010, 70, 1788–1798. [Google Scholar]
- Zhao, Z.; Ma, W.; Zeng, G.; Qi, D.; Ou, L.; Liang, Y. Preoperative serum levels of early prostate cancer antigen (EPCA) predict prostate cancer progression in patients undergoing radical prostatectomy. Prostate 2012, 72, 270–279. [Google Scholar]
- Uetsuki, H.; Tsunemori, H.; Taoka, R.; Haba, R.; Ishikawa, M.; Kakehi, Y. Expression of a novel biomarker, EPCA, in adenocarcinomas and precancerous lesions in the prostate. J. Urol 2005, 174, 514–518. [Google Scholar]
- Sakata, T.; Ferdous, G.; Tsuruta, T.; Satoh, T.; Baba, S.; Muto, T.; Ueno, A.; Kanai, Y.; Endou, H.; Okayasu, I. l-type amino-acid transporter 1 as a novel biomarker for high-grade malignancy in prostate cancer. Pathol. Int 2009, 59, 7–18. [Google Scholar]
- Kanai, Y.; Segawa, H.; Miyamoto, K.; Uchino, H.; Takeda, E.; Endou, H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J. Biol. Chem 1998, 273, 23629–23632. [Google Scholar]
- Kanai, Y.; Endou, H. Heterodimeric amino acid transporters: Molecular biology and pathological and pharmacological relevance. Curr. Drug Metabol 2001, 2, 339–354. [Google Scholar]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar]
- Xie, C.; Kim, H.J.; Haw, J.G.; Kalbasi, A.; Gardner, B.K.; Li, G.; Rao, J.; Chia, D.; Liong, M.; Punzalan, R.R.; et al. A novel multiplex assay combining autoantibodies plus PSA has potential implications for classification of prostate cancer from non-malignant cases. J. Transl. Med 2011, 9, 43. [Google Scholar]
- Ramirez, M.L.; Nelson, E.C.; Evans, C.P. Beyond prostate-specific antigen: Alternate serum markers. Prostate Cancer Prostatic Dis 2008, 11, 216–229. [Google Scholar]
- Beckett, M.L.; Cazares, L.H.; Vlahou, A.; Schellhammer, P.F.; Wright, G.L., Jr. Prostate-specific membrane antigen levels in sera from healthy men and patients with benign prostate hyperplasia or prostate cancer. Clin. Cancer Res. 1999, 5, 4034–4040. [Google Scholar]
- Reiter, R.E.; Gu, Z.; Watabe, T.; Thomas, G.; Szigeti, K.; Davis, E.; Wahl, M.; Nisitani, S.; Yamashiro, J.; Le Beau, M.M.; et al. Prostate stem cell antigen: A cell surface marker overexpressed in prostate cancer. Proc. Natl. Acad. Sci. USA 1998, 95, 1735–1740. [Google Scholar]
- Haese, A.; Graefen, M.; Steuber, T.; Becker, C.; Noldus, J.; Erbersdobler, A.; Huland, E.; Huland, H.; Lilja, H. Total and Gleason grade 4/5 cancer volumes are major contributors of human kallikrein 2, whereas free prostate specific antigen is largely contributed by benign gland volume in serum from patients with prostate cancer or benign prostatic biopsies. J. Urol 2003, 170, 2269–2273. [Google Scholar]
- Kohli, M.; Rothberg, P.G.; Feng, C.; Messing, E.; Joseph, J.; Rao, S.S.; Hendershot, A.; Sahsrabudhe, D. Exploratory study of a KLK2 polymorphism as a prognostic marker in prostate cancer. Cancer Biomark 2010, 7, 101–108. [Google Scholar]
- Xia, C.; Ma, W.; Wang, F.; Hua, S.; Liu, M. Identification of a prostate-specific G-protein coupled receptor in prostate cancer. Oncogene 2001, 20, 5903–5907. [Google Scholar]
- Adley, B.P.; Yang, X.J. Application of alpha-methylacyl coenzyme A racemase immunohistochemistry in the diagnosis of prostate cancer: A review. Anal. Quant. Cytol. Histol 2006, 28, 1–13. [Google Scholar]
- Edwards, S.M.; Evans, D.G.; Hope, Q.; Norman, A.R.; Barbachano, Y.; Bullock, S.; Kote-Jarai, Z.; Meitz, J.; Falconer, A.; Osin, P.; et al. Prostate cancer in BRCA2 germline mutation carriers is associated with poorer prognosis. Br. J. Cancer 2010, 103, 918–924. [Google Scholar]
- Castro, E.; Goh, C.; Olmos, D.; Saunders, E.; Leongamornlert, D.; Tymrakiewicz, M.; Mahmud, N.; Dadaev, T.; Govindasami, K.; Guy, M.; et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J. Clin. Oncol. 2013. [Google Scholar] [CrossRef]
- Vesprini, D.; Liu, S.; Nam, R. Predicting high risk disease using serum and DNA biomarkers. Curr. Opin. Urol 2013, 23, 252–260. [Google Scholar]
- Qin, J.; Wu, S.P.; Creighton, C.J.; Dai, F.; Xie, X.; Cheng, C.M.; Frolov, A.; Ayala, G.; Lin, X.; Feng, X.H.; et al. COUP-TFII inhibits TGF-beta-induced growth barrier to promote prostate tumorigenesis. Nature 2013, 493, 236–240. [Google Scholar]
- Schutzman, J.L.; Martin, G.R. Sprouty genes function in suppression of prostate tumorigenesis. Proc. Natl. Acad. Sci. USA 2012, 109, 20023–20028. [Google Scholar]
- Sowalsky, A.G.; Ye, H.; Bubley, G.J.; Balk, S.P. Clonal progression of prostate cancers from Gleason grade 3 to grade 4. Cancer Res 2013, 73, 1050–1055. [Google Scholar]
- Liong, M.L.; Lim, C.R.; Yang, H.; Chao, S.; Bong, C.W.; Leong, W.S.; Das, P.K.; Loh, C.S.; Lau, B.E.; Yu, C.G.; et al. Blood-based biomarkers of aggressive prostate cancer. PLoS One 2012. [Google Scholar] [CrossRef]
- Voutsadakis, I.A.; Vlachostergios, P.J.; Daliani, D.D.; Karasavvidou, F.; Kakkas, G.; Moutzouris, G.; Melekos, M.D.; Papandreou, C.N. CD10 is inversely associated with nuclear factor-kappa B and predicts biochemical recurrence after radical prostatectomy. Urol. Int 2012, 88, 158–164. [Google Scholar]
- Fleischmann, A.; Rocha, C.; Saxer-Sekulic, N.; Zlobec, I.; Sauter, G.; Thalmann, G.N. High CD10 expression in lymph node metastases from surgically treated prostate cancer independently predicts early death. Virchows Arch 2011, 458, 741–748. [Google Scholar]
- Fleischmann, A.; Schlomm, T.; Huland, H.; Kollermann, J.; Simon, P.; Mirlacher, M.; Salomon, G.; Chun, F.H.; Steuber, T.; Simon, R.; et al. Distinct subcellular expression patterns of neutral endopeptidase (CD10) in prostate cancer predict diverging clinical courses in surgically treated patients. Clin. Cancer Res 2008, 14, 7838–7842. [Google Scholar]
- Xu, L.L.; Sun, C.; Petrovics, G.; Makarem, M.; Furusato, B.; Zhang, W.; Sesterhenn, I.A.; McLeod, D.G.; Sun, L.; Moul, J.W.; et al. Quantitative expression profile of PSGR in prostate cancer. Prostate Cancer Prostatic Dis 2006, 9, 56–61. [Google Scholar]
- Romero, D.; O’Neill, C.; Terzic, A.; Contois, L.; Young, K.; Conley, B.A.; Bergan, R.C.; Brooks, P.C.; Vary, C.P. Endoglin regulates cancer-stromal cell interactions in prostate tumors. Cancer Res 2011, 71, 3482–3493. [Google Scholar]
- Svatek, R.S.; Karam, J.A.; Roehrborn, C.G.; Karakiewicz, P.I.; Slawin, K.M.; Shariat, S.F. Preoperative plasma endoglin levels predict biochemical progression after radical prostatectomy. Clin. Cancer Res 2008, 14, 3362–3366. [Google Scholar]
- Fujita, K.; Ewing, C.M.; Chan, D.Y.; Mangold, L.A.; Partin, A.W.; Isaacs, W.B.; Pavlovich, C.P. Endoglin (CD105) as a urinary and serum marker of prostate cancer. Int. J. Cancer 2009, 124, 664–669. [Google Scholar]
- Pircher, A.; Hilbe, W.; Heidegger, I.; Drevs, J.; Tichelli, A.; Medinger, M. Biomarkers in tumor angiogenesis and anti-angiogenic therapy. Int. J. Mol. Sci 2011, 12, 7077–7099. [Google Scholar]
- Feneley, M.R.; Jan, H.; Granowska, M.; Mather, S.J.; Ellison, D.; Glass, J.; Coptcoat, M.; Kirby, R.S.; Ogden, C.; Oliver, R.T.; et al. Imaging with prostate-specific membrane antigen (PSMA) in prostate cancer. Prostate Cancer Prostatic Dis 2000, 3, 47–52. [Google Scholar]
- Ghosh, A.; Heston, W.D. Tumor target prostate specific membrane antigen (PSMA) and its regulation in prostate cancer. J. Cell Biochem 2004, 91, 528–539. [Google Scholar]
- Freeman, M.R.; Yang, W.; Di Vizio, D. Caveolin-1 and prostate cancer progression. Adv. Exp. Med. Biol 2012, 729, 95–110. [Google Scholar]
- Gumulec, J.; Sochor, J.; Hlavna, M.; Sztalmachova, M.; Krizkova, S.; Babula, P.; Hrabec, R.; Rovny, A.; Adam, V.; Eckschlager, T.; et al. Caveolin-1 as a potential high-risk prostate cancer biomarker. Oncol. Rep 2012, 27, 831–841. [Google Scholar]
- Guo, Y.; Xu, F.; Lu, T.; Duan, Z.; Zhang, Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat. Rev 2012, 7, 904–910. [Google Scholar]
- Han, Z.D.; Bi, X.C.; Qin, W.J.; He, H.C.; Dai, Q.S.; Zou, J.; Ye, Y.K.; Liang, Y.X.; Zeng, G.H.; Chen, Z.N.; et al. CD147 expression indicates unfavourable prognosis in prostate cancer. Pathol. Oncol. Res 2009, 15, 369–374. [Google Scholar]
- Zhong, W.D.; Liang, Y.X.; Lin, S.X.; Li, L.; He, H.C.; Bi, X.C.; Han, Z.D.; Dai, Q.S.; Ye, Y.K.; Chen, Q.B.; et al. Expression of CD147 is associated with prostate cancer progression. Int. J. Cancer 2012, 130, 300–308. [Google Scholar]
- Pertega-Gomes, N.; Vizcaino, J.R.; Miranda-Goncalves, V.; Pinheiro, C.; Silva, J.; Pereira, H.; Monteiro, P.; Henrique, R.M.; Reis, R.M.; Lopes, C.; et al. Monocarboxylate transporter 4 (MCT4) and CD147 overexpression is associated with poor prognosis in prostate cancer. BMC Cancer 2011, 11. [Google Scholar] [CrossRef] [Green Version]
- Donato, R. Intracellular and extracellular roles of S100 proteins. Microsc. Res. Tech 2003, 60, 540–551. [Google Scholar]
- Gupta, S.; Hussain, T.; MacLennan, G.T.; Fu, P.; Patel, J.; Mukhtar, H. Differential expression of S100A2 and S100A4 during progression of human prostate adenocarcinoma. J. Clin. Oncol 2003, 21, 106–112. [Google Scholar]
- Rehman, I.; Azzouzi, A.R.; Cross, S.S.; Deloulme, J.C.; Catto, J.W.; Wylde, N.; Larre, S.; Champigneuille, J.; Hamdy, F.C. Dysregulated expression of S100A11 (calgizzarin) in prostate cancer and precursor lesions. Hum Pathol 2004, 35, 1385–1391. [Google Scholar]
- Rehman, I.; Cross, S.S.; Catto, J.W.; Leiblich, A.; Mukherjee, A.; Azzouzi, A.R.; Leung, H.Y.; Hamdy, F.C. Promoter hyper-methylation of calcium binding proteins S100A6 and S100A2 in human prostate cancer. Prostate 2005, 65, 322–330. [Google Scholar]
- Hermani, A.; Hess, J.; De Servi, B.; Medunjanin, S.; Grobholz, R.; Trojan, L.; Angel, P.; Mayer, D. Calcium-binding proteins S100A8 and S100A9 as novel diagnostic markers in human prostate cancer. Clin. Cancer Res 2005, 11, 5146–5152. [Google Scholar]
- Kollermann, J.; Schlomm, T.; Bang, H.; Schwall, G.P.; von Eichel-Streiber, C.; Simon, R.; Schostak, M.; Huland, H.; Berg, W.; Sauter, G.; et al. Expression and prognostic relevance of annexin A3 in prostate cancer. Eur. Urol 2008, 54, 1314–1323. [Google Scholar]
- Leman, E.S.; Getzenberg, R.H. Biomarkers for prostate cancer. J. Cell Biochem 2009, 108, 3–9. [Google Scholar]
- Gerke, V.; Creutz, C.E.; Moss, S.E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol 2005, 6, 449–461. [Google Scholar]
- Pisitkun, T.; Shen, R.F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar]
- Schostak, M.; Schwall, G.P.; Poznanovic, S.; Groebe, K.; Muller, M.; Messinger, D.; Miller, K.; Krause, H.; Pelzer, A.; Horninger, W.; et al. Annexin A3 in urine: A highly specific noninvasive marker for prostate cancer early detection. J. Urol 2009, 181, 343–353. [Google Scholar]
- Kattan, M.W.; Shariat, S.F.; Andrews, B.; Zhu, K.; Canto, E.; Matsumoto, K.; Muramoto, M.; Scardino, P.T.; Ohori, M.; Wheeler, T.M.; et al. The addition of interleukin-6 soluble receptor and transforming growth factor beta1 improves a preoperative nomogram for predicting biochemical progression in patients with clinically localized prostate cancer. J. Clin. Oncol 2003, 21, 3573–3579. [Google Scholar]
- Shariat, S.F.; Shalev, M.; Menesses-Diaz, A.; Kim, I.Y.; Kattan, M.W.; Wheeler, T.M.; Slawin, K.M. Preoperative plasma levels of transforming growth factor beta(1) (TGF-beta(1)) strongly predict progression in patients undergoing radical prostatectomy. J. Clin. Oncol 2001, 19, 2856–2864. [Google Scholar]
- Morton, D.M.; Barrack, E.R. Modulation of transforming growth factor beta 1 effects on prostate cancer cell proliferation by growth factors and extracellular matrix. Cancer Res 1995, 55, 2596–2602. [Google Scholar]
- Shariat, S.F.; Kattan, M.W.; Traxel, E.; Andrews, B.; Zhu, K.; Wheeler, T.M.; Slawin, K.M. Association of pre- and postoperative plasma levels of transforming growth factor beta(1) and interleukin 6 and its soluble receptor with prostate cancer progression. Clin. Cancer Res 2004, 10, 1992–1999. [Google Scholar]
- Avgeris, M.; Mavridis, K.; Scorilas, A. Kallikrein-related peptidases in prostate, breast, and ovarian cancers: From pathobiology to clinical relevance. Biol. Chem 2012, 393, 301–317. [Google Scholar]
- Gallagher, D.J.; Vijai, J.; Cronin, A.M.; Bhatia, J.; Vickers, A.J.; Gaudet, M.M.; Fine, S.; Reuter, V.; Scher, H.I.; Hallden, C.; et al. Susceptibility loci associated with prostate cancer progression and mortality. Clin. Cancer Res 2010, 16, 2819–2832. [Google Scholar]
- Harries, L.W.; Perry, J.R.; McCullagh, P.; Crundwell, M. Alterations in LMTK2, MSMB and HNF1B gene expression are associated with the development of prostate cancer. BMC Cancer 2010, 10. [Google Scholar] [CrossRef] [Green Version]
- FitzGerald, L.M.; Zhang, X.; Kolb, S.; Kwon, E.M.; Liew, Y.C.; Hurtado-Coll, A.; Knudsen, B.S.; Ostrander, E.A.; Stanford, J.L. Investigation of the relationship between prostate cancer and MSMB and NCOA4 genetic variants and protein expression. Hum. Mutat 2013, 34, 149–156. [Google Scholar]
- Huang, P.Y.; Best, O.G.; Belov, L.; Mulligan, S.P.; Christopherson, R.I. Surface profiles for subclassification of chronic lymphocytic leukemia. Leuk. Lymphoma 2012, 53, 1046–1056. [Google Scholar]
- Choudhury, A.D.; Eeles, R.; Freedland, S.J.; Isaacs, W.B.; Pomerantz, M.M.; Schalken, J.A.; Tammela, T.L.; Visakorpi, T. The role of genetic markers in the management of prostate cancer. Eur. Urol 2012, 4, 577–587. [Google Scholar]
- Eeles, R.A.; Olama, A.A.; Benlloch, S.; Saunders, E.J.; Leongamornlert, D.A.; Tymrakiewicz, M.; Ghoussaini, M.; Luccarini, C.; Dennis, J.; Jugurnauth-Little, S.; et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet 2013, 45, 385–391. [Google Scholar]
- Salagierski, M.; Schalken, J.A. Molecular diagnosis of prostate cancer: PCA3 and TMPRSS2:ERG gene fusion. J. Urol 2012, 187, 795–801. [Google Scholar]
- Van Gils, M.P.; Hessels, D.; van Hooij, O.; Jannink, S.A.; Peelen, W.P.; Hanssen, S.L.; Witjes, J.A.; Cornel, E.B.; Karthaus, H.F.; Smits, G.A.; et al. The time-resolved fluorescence-based PCA3 test on urinary sediments after digital rectal examination; a Dutch multicenter validation of the diagnostic performance. Clin. Cancer Res 2007, 13, 939–943. [Google Scholar]
- King, J.C.; Xu, J.; Wongvipat, J.; Hieronymus, H.; Carver, B.S.; Leung, D.H.; Taylor, B.S.; Sander, C.; Cardiff, R.D.; Couto, S.S.; et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat. Genet 2009, 41, 524–526. [Google Scholar]
- Hessels, D.; Smit, F.P.; Verhaegh, G.W.; Witjes, J.A.; Cornel, E.B.; Schalken, J.A. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin. Cancer Res 2007, 13, 5103–5108. [Google Scholar]
- Salami, S.S.; Schmidt, F.; Laxman, B.; Regan, M.M.; Rickman, D.S.; Scherr, D.; Bueti, G.; Siddiqui, J.; Tomlins, S.A.; Wei, J.T.; et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol. Oncol. 2011. [Google Scholar] [CrossRef]
- Saramaki, O.R.; Harjula, A.E.; Martikainen, P.M.; Vessella, R.L.; Tammela, T.L.; Visakorpi, T. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin. Cancer Res 2008, 14, 3395–3400. [Google Scholar]
- De Muga, S.; Hernandez, S.; Salido, M.; Lorenzo, M.; Agell, L.; Juanpere, N.; Lorente, J.A.; Serrano, S.; Lloreta, J. CXCR4 mRNA overexpression in high grade prostate tumors: Lack of association with TMPRSS2-ERG rearrangement. Cancer Biomark 2012, 12, 21–30. [Google Scholar]
- Du Pasquier, L. Innate immunity in early chordates and the appearance of adaptive immunity. Comptes Rendus Biol 2004, 327, 591–601. [Google Scholar]
- Yeh, J.H.; Sidhu, S.S.; Chan, A.C. Regulation of a late phase of T cell polarity and effector functions by Crtam. Cell 2008, 132, 846–859. [Google Scholar]
- Roth, C.; Schuierer, M.; Gunther, K.; Buettner, R. Genomic structure and DNA binding properties of the human zinc finger transcriptional repressor AP-2rep (KLF12). Genomics 2000, 63, 384–390. [Google Scholar]
- Catto, J.W.; Alcaraz, A.; Bjartell, A.S.; De Vere White, R.; Evans, C.P.; Fussel, S.; Hamdy, F.C.; Kallioniemi, O.; Mengual, L.; Schlomm, T.; et al. MicroRNA in prostate, bladder, and kidney cancer: A systematic review. Eur. Urol 2011, 59, 671–681. [Google Scholar]
- Catto, J.W.; Miah, S.; Owen, H.C.; Bryant, H.; Myers, K.; Dudziec, E.; Larre, S.; Milo, M.; Rehman, I.; Rosario, D.J.; et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res 2009, 69, 8472–8481. [Google Scholar]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar]
- Ilic, D.; Neuberger, M.M.; Djulbegovic, M.; Dahm, P. Screening for prostate cancer. Cochrane Database Syst. Rev. 2013. [Google Scholar] [CrossRef]
- Nikitina, E.G.; Urazova, L.N.; Stegny, V.N. MicroRNAs and human cancer. Exp. Oncol 2012, 34, 2–8. [Google Scholar]
- Sun, T.; Wang, Q.; Balk, S.; Brown, M.; Lee, G.S.; Kantoff, P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res 2009, 69, 3356–3363. [Google Scholar]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar]
- Dabir, P.D.; Ottosen, P.; Hoyer, S.; Hamilton-Dutoit, S. Comparative analysis of three- and two-antibody cocktails to AMACR and basal cell markers for the immunohistochemical diagnosis of prostate carcinoma. Diagn. Pathol. 2012, 7. [Google Scholar] [CrossRef]
- Shariat, S.F.; Scherr, D.S.; Gupta, A.; Bianco, F.J., Jr; Karakiewicz, P.I.; Zeltser, I.S.; Samadi, D.B.; Akhavan, A. Emerging biomarkers for prostate cancer diagnosis, staging, and prognosis. Arch. Esp. Urol. 2011, 64, 681–694. [Google Scholar]
- Jiang, Z.; Fanger, G.R.; Woda, B.A.; Banner, B.F.; Algate, P.; Dresser, K.; Xu, J.; Chu, P.G. Expression of alpha-methylacyl-CoA racemase (P504s) in various malignant neoplasms and normal tissues: Astudy of 761 cases. Hum. Pathol 2003, 34, 792–796. [Google Scholar]
- Sreekumar, A.; Laxman, B.; Rhodes, D.R.; Bhagavathula, S.; Harwood, J.; Giacherio, D.; Ghosh, D.; Sanda, M.G.; Rubin, M.A.; Chinnaiyan, A.M. Humoral immune response to alpha-methylacyl-CoA racemase and prostate cancer. J. Natl. Cancer Inst 2004, 96, 834–843. [Google Scholar]
- Prensner, J.R.; Rubin, M.A.; Wei, J.T.; Chinnaiyan, A.M. Beyond PSA: The next generation of prostate cancer biomarkers. Sci. Transl. Med. 2012, 4. [Google Scholar] [CrossRef]
- Ouyang, B.; Leung, Y.K.; Wang, V.; Chung, E.; Levin, L.; Bracken, B.; Cheng, L.; Ho, S.M. Alpha-Methylacyl-CoA racemase spliced variants and their expression in normal and malignant prostate tissues. Urology 2011, 77, 241–247. [Google Scholar]
- Lloyd, M.D.; Yevglevskis, M.; Lee, G.L.; Wood, P.J.; Threadgill, M.D.; Woodman, T.J. Alpha-Methylacyl-CoA racemase (AMACR): Metabolic enzyme, drug metabolizer and cancer marker P504S. Prog. Lipid Res 2013, 52, 220–230. [Google Scholar]
- Rubin, M.A.; Bismar, T.A.; Andren, O.; Mucci, L.; Kim, R.; Shen, R.; Ghosh, D.; Wei, J.T.; Chinnaiyan, A.M.; Adami, H.O.; et al. Decreased alpha-methylacyl CoA racemase expression in localized prostate cancer is associated with an increased rate of biochemical recurrence and cancer-specific death. Cancer Epidemiol. Biomark. Prev 2005, 14, 1424–1432. [Google Scholar]
- Saedi, M.S.; Zhu, Z.; Marker, K.; Liu, R.S.; Carpenter, P.M.; Rittenhouse, H.; Mikolajczyk, S.D. Human kallikrein 2 (hK2), but not prostate-specific antigen (PSA), rapidly complexes with protease inhibitor 6 (PI-6) released from prostate carcinoma cells. Int. J. Cancer 2001, 94, 558–563. [Google Scholar]
- Potter, S.R.; Partin, A.W. Tumor markers: An update on human kallikrein 2. Rev. Urol 2000, 2, 221–222. [Google Scholar]
- Recker, F.; Kwiatkowski, M.K.; Piironen, T.; Pettersson, K.; Huber, A.; Lummen, G.; Tscholl, R. Human glandular kallikrein as a tool to improve discrimination of poorly differentiated and non-organ-confined prostate cancer compared with prostate-specific antigen. Urology 2000, 55, 481–485. [Google Scholar]
- Steuber, T.; Vickers, A.J.; Haese, A.; Becker, C.; Pettersson, K.; Chun, F.K.; Kattan, M.W.; Eastham, J.A.; Scardino, P.T.; Huland, H.; et al. Risk assessment for biochemical recurrence prior to radical prostatectomy: Significant enhancement contributed by human glandular kallikrein 2 (hK2) and free prostate specific antigen (PSA) in men with moderate PSA-elevation in serum. Int. J. Cancer 2006, 118, 1234–1240. [Google Scholar]
- Morgan, R.; Boxall, A.; Bhatt, A.; Bailey, M.; Hindley, R.; Langley, S.; Whitaker, H.C.; Neal, D.E.; Ismail, M.; Whitaker, H.; et al. Engrailed-2 (EN2): A tumor specific urinary biomarker for the early diagnosis of prostate cancer. Clin. Cancer Res 2011, 17, 1090–1098. [Google Scholar]
- Bose, S.K.; Bullard, R.S.; Donald, C.D. Oncogenic role of engrailed-2 (en-2) in prostate cancer cell growth and survival. Transl. Oncogenomics 2008, 3, 37–43. [Google Scholar]
- Pandha, H.; Sorensen, K.D.; Orntoft, T.F.; Langley, S.; Hoyer, S.; Borre, M.; Morgan, R. Urinary engrailed-2 (EN2) levels predict tumour volume in men undergoing radical prostatectomy for prostate cancer. BJU Int 2012, 110, 287–292. [Google Scholar]
- Launay, G.; Teletchea, S.; Wade, F.; Pajot-Augy, E.; Gibrat, J.F.; Sanz, G. Automatic modeling of mammalian olfactory receptors and docking of odorants. Protein Eng. Des. Sel 2012, 25, 377–386. [Google Scholar]
- Wang, X.; Yu, J.; Sreekumar, A.; Varambally, S.; Shen, R.; Giacherio, D.; Mehra, R.; Montie, J.E.; Pienta, K.J.; Sanda, M.G.; et al. Autoantibody signatures in prostate cancer. N. Engl. J. Med 2005, 353, 1224–1235. [Google Scholar]
- Massoner, P.; Lueking, A.; Goehler, H.; Hopfner, A.; Kowald, A.; Kugler, K.G.; Amersdorfer, P.; Horninger, W.; Bartsch, G.; Schulz-Knappe, P.; et al. Serum-autoantibodies for discovery of prostate cancer specific biomarkers. Prostate 2012, 72, 427–436. [Google Scholar]
- Barber, N.; Gez, S.; Belov, L.; Mulligan, S.P.; Woolfson, A.; Christopherson, R.I. Profiling CD antigens on leukaemias with an antibody microarray. FEBS Lett 2009, 583, 1785–1791. [Google Scholar]
- Belov, L.; Huang, P.; Chrisp, J.S.; Mulligan, S.P.; Christopherson, R.I. Screening microarrays of novel monoclonal antibodies for binding to T-, B- and myeloid leukaemia cells. J. Immunol. Methods 2005, 305, 10–19. [Google Scholar]
- Kaufman, K.L.; Belov, L.; Huang, P.; Mactier, S.; Scolyer, R.A.; Mann, G.J.; Christopherson, R.I. An extended antibody microarray for surface profiling metastatic melanoma. J. Immunol. Methods 2010, 358, 23–34. [Google Scholar]
- Lal, S.; Brown, A.; Nguyen, L.; Braet, F.; Dyer, W.; Dos Remedios, C. Using antibody arrays to detect microparticles from acute coronary syndrome patients based on cluster of differentiation (CD) antigen expression. Mol. Cell Proteomics 2009, 8, 799–804. [Google Scholar]
- Zhou, J.; Belov, L.; Huang, P.Y.; Shin, J.S.; Solomon, M.J.; Chapuis, P.H.; Bokey, L.; Chan, C.; Clarke, C.; Clarke, S.J.; et al. Surface antigen profiling of colorectal cancer using antibody microarrays with fluorescence multiplexing. J. Immunol. Methods 2010, 355, 40–51. [Google Scholar]
- Wu, J.Q.; Wang, B.; Belov, L.; Chrisp, J.; Learmont, J.; Dyer, W.B.; Zaunders, J.; Cunningham, A.L.; Dwyer, D.E.; Saksena, N.K. Antibody microarray analysis of cell surface antigens on CD4+ and CD8+ T cells from HIV+ individuals correlates with disease stages. Retrovirology 2007, 4. [Google Scholar] [CrossRef]
- Jiao, J.; Hindoyan, A.; Wang, S.; Tran, L.M.; Goldstein, A.S.; Lawson, D.; Chen, D.; Li, Y.; Guo, C.; Zhang, B.; et al. Identification of CD166 as a Surface Marker for Enriching Prostate Stem/Progenitor and Cancer Initiating Cells. PLoS One 2012, 7. [Google Scholar] [CrossRef]
- Hao, J.; Madigan, M.C.; Khatri, A.; Power, C.A.; Hung, T.T.; Beretov, J.; Chang, L.; Xiao, W.; Cozzi, P.J.; Graham, P.H.; et al. In Vitro and In Vivo Prostate Cancer Metastasis and Chemoresistance Can Be Modulated by Expression of either CD44 or CD147. PLoS One 2012, 7. [Google Scholar] [CrossRef]
- Gyorgy, B.; Szabo, T.G.; Pasztoi, M.; Pal, Z.; Misjak, P.; Aradi, B.; Laszlo, V.; Pallinger, E.; Pap, E.; Kittel, A.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell Mol. Life Sci 2011, 68, 2667–2688. [Google Scholar]
- Duijvesz, D.; Luider, T.; Bangma, C.H.; Jenster, G. Exosomes as biomarker treasure chests for prostate cancer. Eur. Urol 2011, 59, 823–831. [Google Scholar]
- Nepple, K.G.; Wahls, T.L.; Hillis, S.L.; Joudi, F.N. Gleason Score and Laterality Concoordance Between Prostate Biopsy and Prostatectomy Specimens. Int. J. Braz. Urol 2009, 35, 559–564. [Google Scholar]
- Yang, C.; Robbins, P.D. The roles of tumor-derived exosomes in cancer pathogenesis. Clin. Dev. Immunol. 2011. [Google Scholar] [CrossRef]
- Yang, Y.; Xiu, F.; Cai, Z.; Wang, J.; Wang, Q.; Fu, Y.; Cao, X. Increased induction of antitumor response by exosomes derived from interleukin-2 gene-modified tumor cells. J. Cancer Res. Clin. Oncol 2007, 133, 389–399. [Google Scholar]
- Ludwig, A.K.; Giebel, B. Exosomes: Small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol 2012, 44, 11–15. [Google Scholar]
- Ristorcelli, E.; Beraud, E.; Verrando, P.; Villard, C.; Lafitte, D.; Sbarra, V.; Lombardo, D.; Verine, A. Human tumor nanoparticles induce apoptosis of pancreatic cancer cells. FASEB 2008, 22, 3358–3369. [Google Scholar]
- Wood, S.L.; Knowles, M.A.; Thompson, D.; Selby, P.J.; Banks, R.E. Proteomic studies of urinary biomarkers for prostate, bladder and kidney cancers. Nat. Rev. Urol 2013, 10, 206–218. [Google Scholar]
- Delcayre, A.; Shu, H.; Le Pecq, J.B. Dendritic cell-derived exosomes in cancer immunotherapy: Exploiting nature’s antigen delivery pathway. Expert Rev. Anticancer Ther 2005, 5, 537–547. [Google Scholar]
- Thery, C.; Boussac, M.; Veron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol 2001, 166, 7309–7318. [Google Scholar]
- Ronquist, G.; Brody, I. The prostasome: Its secretion and function in man. Biochim. Biophys. Acta 1985, 822, 203–218. [Google Scholar]
- Ronquist, G.; Nilsson, B.O. The Janus-faced nature of prostasomes: Their pluripotency favours the normal reproductive process and malignant prostate growth. Prostate Cancer Prostatic Dis 2004, 7, 21–31. [Google Scholar]
- Tavoosidana, G.; Ronquist, G.; Darmanis, S.; Yan, J.; Carlsson, L.; Wu, D.; Conze, T.; Ek, P.; Semjonow, A.; Eltze, E.; et al. Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 8809–8814. [Google Scholar]
- Saez, F.; Frenette, G.; Sullivan, R. Epididymosomes and prostasomes: Their roles in posttesticular maturation of the sperm cells. J. Androl 2003, 24, 149–154. [Google Scholar]
- Oon, S.F.; Pennington, S.R.; Fitzpatrick, J.M.; Watson, R.W. Biomarker research in prostate cancer—Towards utility, not futility. Nat. Rev. Urol 2011, 8, 131–138. [Google Scholar]
- Ronquist, G. Prostasomes are mediators of intercellular communication: From basic research to clinical implications. J. Int. Med 2012, 271, 400–413. [Google Scholar]
- Sahlen, G.E.; Egevad, L.; Ahlander, A.; Norlen, B.J.; Ronquist, G.; Nilsson, B.O. Ultrastructure of the secretion of prostasomes from benign and malignant epithelial cells in the prostate. Prostate 2002, 53, 192–199. [Google Scholar]
- Olmos, D.; Baird, R.D.; Yap, T.A.; Massard, C.; Pope, L.; Sandhu, S.K.; Attard, G.; Dukes, J.; Papadatos-Pastos, D.; Grainger, P.; et al. Baseline circulating tumor cell counts significantly enhance a prognostic score for patients participating in phase I oncology trials. Clin. Cancer Res 2011, 17, 5188–5196. [Google Scholar]
- Attard, G.; de Bono, J.S. Utilizing circulating tumor cells: Challenges and pitfalls. Curr. Opin. Genet. Dev 2011, 21, 50–58. [Google Scholar]
- Shaffer, D.R.; Leversha, M.A.; Danila, D.C.; Lin, O.; Gonzalez-Espinoza, R.; Gu, B.; Anand, A.; Smith, K.; Maslak, P.; Doyle, G.V.; et al. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clin. Cancer Res 2007, 13, 2023–2029. [Google Scholar]
- Bednarz-Knoll, N.; Alix-Panabieres, C.; Pantel, K. Clinical relevance and biology of circulating tumor cells. Breast Cancer Res. 2011, 13. [Google Scholar] [CrossRef]
- Laxman, B.; Morris, D.S.; Yu, J.; Siddiqui, J.; Cao, J.; Mehra, R.; Lonigro, R.J.; Tsodikov, A.; Wei, J.T.; Tomlins, S.A.; et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res 2008, 68, 645–649. [Google Scholar]
| Marker | Product | Biological Function/Relation to PCa | Reference |
|---|---|---|---|
| B7-H3 (CD276) | Co-stimulatory molecule | May act as antigen-specific inhibitor of T-cell-mediated anti-tumoral immunity. Increased expression worsens PCa prognosis. | [50–53] |
| Ki-67 | Nuclear protein | Cell-cycle-proliferation marker. Possibly a prolific predictive marker for men with low grade, low volume PCa after radical prostatectomy. Associated with metastasis and survival outcome. | [54,55] |
| EPCA | Early Prostate Cancer Antigen Nuclear matrix protein | PCa-associated nuclear structural protein measured in serum. Expressed in prostate adenocarcinoma and benign tissue; correlation with tumor progression and poor prognosis. | [56–58] |
| LAT1 (CD98) | Amino acid transporter | Primary function is to transport essential amino acids. Elevated LAT1 expression in PCa proposed as a novel independent biomarker of high-grade malignancy. LAT1 activity is considered essential for cancer cell proliferation. | [59–61] |
| PCA3 | Non-coding RNA | Produced in the prostate. Overexpressed compared to non-malignant prostate tissue with a high specificity. | [2,62,63] |
| PSCA | Prostate Stem Cell Antigen, a membrane glycoprotein | Involved in the regulation of cell proliferation. Up-regulated in the majority of PCas however, exact biological function is unknown. Increased expression is associated with Gleason score, seminal vesicle invasion, and capsular invasion in PCa. | [55,64–66] |
| TMPRSS2-ERG gene fusion | Transcription factor | Secreted from prostate epithelial cells; expressed in malignant prostate tissue. Independent marker of disease progression and known marker of poor prognosis. Detected in urine; small-scale studies suggest high specificity and sensitivity. | [31,32,67–70] |
| BRCA1/BRCA2 | Tumor suppressor | Both BRCA1 and BRCA2 are involved in maintaining genome stability as members of the ATM/ATR CHK2 DNA damage repair pathway. BRCA2 is associated with aggressive tumors and poor survival outcome. BRCA2 has prognostic ability however further experimental data is needed for BRCA1. | [71–73] |
| PTEN | Phosphatase and Tensin homologue; protein phosphatase | Tumor suppressor involved in modulating the PI3-K/AKT signaling pathway. PTEN inactivating mutations/deletion occur in many tumors and result in rapid cell growth and division. It is associated with severe tumor stage; however, PTEN is not PCa specific It is among one of the most frequent genetic inactivation’s present in PCa. | [74–76] |
| PI3K | Phosphoinositide-3-kinase; Protein kinase. | One of the most common genomic alterations in human PCa contributing to cellular transformation and cancer development. Possibly a key mechanism supporting progression toward androgen-independent PCa. | [74,75] |
| PCa 7 gene panel CTAM, CXCR3, FCRL3, KIAA1143, KLF12, TMEM204,SAMSN1 | Uncharacterized, Chemokine receptor 3, Fc receptor-like 3, uncharacterized, Kruppel-like factor 12, transmembrane protein 204, and SH3 domain and nuclear localization signals 1 respectively | A panel of 7 genes derived from blood mRNA could distinguish between aggressive PCa and healthy patients with a high sensitivity (83%) and specificity (80%). Genes involved in regulating the immune response and gene transcription regulation in oncogenesis. | [77] |
| MME | Membrane metallo-endopeptidase/CD10 | Inactivates several peptide hormones including glucagon, abundant in the kidney. Candidate cancer biomarker associated with PCa progression. A low level of CD10 is a possible prognostic indicator for biochemical relapse and early death as a result of lymph node metastases. Additionally may aid in personalized patient treatment/management however this marker needs to be further validated. | [78–80] |
| PSGR | Prostate Specific G protein-coupled receptor Protein-olfactory receptor | Increased PSGR expression is associated with PCa progression compared to normal tissue, possibly involved in cell proliferation. Significant PSGR alterations are observed in primary PCa cases and overexpression is associated with higher pathological stage. | [69,81] |
| Protein Marker | Protein type | Biological Function/Relation to PCa | Reference |
|---|---|---|---|
| Alpha-methylacyl-CoA Racemase (AMACR) | Racemase | Metabolize fatty acids in the body. Over-expressed in PCa tissue; detected with a high sensitivity and specificity in blood and urine. | [74–80] |
| Endoglin (CD105) | Trans membrane glycoprotein | Expressed by human vascular endothelial cells thought to play a pivotal role in endothelial cell proliferation. Elevated in prostatic fluid of men with large volume PCa. | [55,82–84] |
| Engrailed 2; (EN-2) | Transcription factor | Involved in early embryonic development and re-expressed by PCa cells. EN-2 detection in urine as a test for diagnosing and detecting PCa. Although further validation is required, it appears it is more reliable than PSA and elevated expression is associated with increased tumor stage. | [57–59] |
| Prostate-specific membrane antigen (PSMA) | Type II integral membrane glycoprotein | Overexpressed on prostate tumor cells and in the neovasculature of most solid prostate tumors, but not in the vasculature of normal tissues. May play an important role in the progression of PCa. | [55,85–87] |
| Caveolin-1 | Integral membrane protein | Mediates aspects of cholesterol and fatty acid metabolism. Circulating levels of serum Caveolin-1 correlate with extent of PCa. | [88,89] |
| Interleukin-6 (IL-6) | Cytokine | Involved in hematopoiesis and mediates B cell differentiation. Clinical studies reveal increased serum IL-6 concentrations in patients are associated with advanced PCa tumor stage. | [55,90] |
| CD147 | Membrane glycoprotein | Over-expressed in many human solid tumors. Involved in tumor invasion and angiogenesis. Increased expression of CD147 is associated with PCa progression and poor prognosis. May serve as an independent predictor of biochemical recurrence and development of PCa metastasis. | [91–93] |
| S100 Protein Family | Calcium-binding-protein family | Expressed in various solid tumors. Detection may be useful for diagnosis, monitoring and possible therapeutic targets. Involved in protein phosphorylation, enzyme activity, calcium homeostasis, and regulation of transcription factors, macrophage activators and modulators of cell proliferation. S100A2, S100A4, S100A8, S100A9 and S100A11 are associated with PCa recurrence and advanced pathological stage. | [94–98] |
| Annexin A3 (ANXA3) | Cell adhesion protein | A calcium and phospholipid binding protein, primarily found in urine. Implicated in cell differentiation, migration and immunomodulation. Increases the specificity and ability of PSA to discriminate between PCa stages. | [99–103] |
| TGF-Beta 1 | Cytokine | Growth factor involved in the regulation of cellular proliferation, immune response and differentiation. Increased expression correlates with severe tumor grade, tumor invasion, PCa metastasis and biochemical recurrence. TGF-Beta needs to be validated before becoming a PCa biomarker. | [74,104–107] |
| Human Kallikrein-2 (KLK2) | Serine protease | Serine protease that is highly expressed in prostate tissue and involved regulating semen liquefaction by activating pro-KLK3 to its active form (PSA), facilitating both tumorigenisis and disease progression to the advanced stages of PCa. Studies have shown a strong correlation with PCa-specific survival however further studies with larger cohorts are needed to confirm these observations. | [68,108,109] |
| Beta-microseminoprotein (MSMB) | Immunoglobulin binding factor | Secreted by epithelial cells of the prostate as well as other major organs. MSMB is a member of the immunoglobulin binding family. Exact function of MSMB is unknown but may have an autocrine (inhibin-like) role. The genetic variant rs10993994 is associated with PCa risk however further investigation is required to evaluate the predictive value of this marker. | [110,111] |
| Category | Summary | Reference |
|---|---|---|
| Circulating tumor cells (CTCs) | CTCs detected in blood have been proposed for monitoring disease progression and evaluating effectiveness of cancer therapy. They carry important information specific to tumor type and stage however low CTC detection in blood proves to be a technical hurdle. Prostate cancer derived CTCs possess those same mutations present in the primary tumor (PTEN, TMPRSS2, AMACR), which may provide a more readily accessible source of important prognostic information for patients. To date the high cost associated with their analysis and controversial clinical relevance has prevented their use in clinical setting. | [175,176] |
| Prostasomes | Prostasomes are sub-micrometer membranous vesicles, generated from normal and malignant prostate cells. They are found in blood, urine, semen and prostatic fluid. An increased abundance of prostasomes have been associated with PCa and elevated Gleason score. They carry specific markers (CD46, CD55, CD59) that play a role in the immune system. Additionally, they carry specific molecules, both intracellular and extracellular, that may be specific to PCa and aid in the discovery of new PCa biomarkers. | [170,173,174] |
| Exosomes | Exosomes are cell-derived vesicles isolated from blood, urine and cell lines. Exosomes are released from most cancer types and possess immunosuppressive properties thought to play a significant role in oncogenesis. All exosomes express specific markers (CD9, Alix, CD81) that enable easier detection and isolation. In addition to these common markers, exosomes express specific markers unique to PCa and the cells from which they were derived. They can potentially characterize different stages of PCa and hold prognostic potential. | [159,161] |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Velonas, V.M.; Woo, H.H.; Remedios, C.G.d.; Assinder, S.J. Current Status of Biomarkers for Prostate Cancer. Int. J. Mol. Sci. 2013, 14, 11034-11060. https://doi.org/10.3390/ijms140611034
Velonas VM, Woo HH, Remedios CGd, Assinder SJ. Current Status of Biomarkers for Prostate Cancer. International Journal of Molecular Sciences. 2013; 14(6):11034-11060. https://doi.org/10.3390/ijms140611034
Chicago/Turabian StyleVelonas, Vicki M., Henry H. Woo, Cristobal G. dos Remedios, and Stephen J. Assinder. 2013. "Current Status of Biomarkers for Prostate Cancer" International Journal of Molecular Sciences 14, no. 6: 11034-11060. https://doi.org/10.3390/ijms140611034




