The Influence of Light Quality, Circadian Rhythm, and Photoperiod on the CBF-Mediated Freezing Tolerance
Abstract
:1. Introduction
2. Cold Acclimation and the CBF Regulon
3. Light Quality and Cold Acclimation
4. Circadian Regulation of Cold Acclimation
5. Photoperiodic Regulation of Cold Acclimation
6. Genetically Engineering Cold-Stress Tolerance in Crop Plants
7. Conclusions
Acknowledgments
Conflict of Interest
References
- Lindlof, A. Interplay between low-temperature pathways and light reduction. Plant Signal. Behav 2010, 5, 820–825. [Google Scholar]
- Levitt, J. Responses of Plants to Environmental Stresses, 2nd ed.; Academic Press: New York, NY, USA, 1980. [Google Scholar]
- Medina, J.; Catala, R.; Salinas, J. The CBFs: Three Arabidopsis transcription factors to cold acclimate. Plant Sci 2011, 180, 3–11. [Google Scholar]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol 1999, 50, 571–599. [Google Scholar]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci 2007, 12, 444–451. [Google Scholar]
- Janska, A.; Marsik, P.; Zelenkova, S.; Ovesna, J. Cold stress and acclimation—What is important for metabolic adjustment? Plant Biol 2010, 12, 395–405. [Google Scholar]
- Gilmour, S.J.; Zarka, D.G.; Stockinger, E.J.; Salazar, M.P.; Houghton, J.M.; Thomashow, M.F. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 1998, 16, 433–442. [Google Scholar]
- Jurczyk, B.; Rapacz, M.; Budzisz, K.; Barcik, W.; Sasal, M. The effects of cold, light and time of day during low-temperature shift on the expression of CBF6, FpCor14b and LOS2 in Festuca pratensis. Plant Sci 2012, 183, 143–148. [Google Scholar]
- Wanner, L.A.; Junttila, O. Cold-induced freezing tolerance in Arabidopsis. Plant Physiol 1999, 120, 391–400. [Google Scholar]
- Crosatti, C.; Polverino de Laureto, P.; Bassi, R.; Cattivelli, L. The interaction between cold and light controls the expression of the cold-regulated barley gene cor14b and the accumulation of the corresponding protein. Plant Physiol 1999, 119, 671–680. [Google Scholar]
- Kim, H.J.; Kim, Y.K.; Park, J.Y.; Kim, J. Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J 2002, 29, 693–704. [Google Scholar]
- Soitamo, A.J.; Piippo, M.; Allahverdiyeva, Y.; Battchikova, N.; Aro, E.M. Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant Biol 2008, 8, 13, :1–13:20.. [Google Scholar]
- Olsen, J.E.; Nilsen, O.J.J.; Eriksson, M.E.; Martinussen, I.; Olsson, O.; Sandberg, G.; Moritz, T. Ectopic expression of oat phytochrome A in hybrid aspen changes critical daylength for growth and prevents cold acclimatization. Plant J 1997, 12, 1339–1350. [Google Scholar]
- Catala, R.; Medina, J.; Salinas, J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 16475–16480. [Google Scholar]
- Dong, M.A.; Farre, E.M.; Thomashow, M.F. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 7241–7246. [Google Scholar]
- Fowler, S.G.; Cook, D.; Thomashow, M.F. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 2005, 137, 961–968. [Google Scholar]
- Harrison, L.C.; Weiser, C.J.; Burke, M.J. Environmental and seasonal factors affecting the frost-induced stage of cold acclimation in Cornus stolonifera Michx. Plant Physiol 1978, 62, 894–898. [Google Scholar]
- Weiser, C.J. Cold Resistance and Injury in Woody Plants: Knowledge of hardy plant adaptations to freezing stress may help us to reduce winter damage. Science 1970, 169, 1269–1278. [Google Scholar]
- Lee, C.M.; Thomashow, M.F. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 15054–15059. [Google Scholar]
- Ruelland, E.; Vaultier, M.-N.; Zachowski, A.; Hurry, V. Cold signaling and cold acclimation in plants. Adv. Bot. Res 2009, 49, 36–54. [Google Scholar]
- Mita, T.; Shibaoka, H. Gibberellin stabilizes microtubules in onion leaf sheath cells. Proteoplasma 1984, 119, 100–109. [Google Scholar]
- Nishida, I.; Murata, N. Chilling sensitivity in plants and cyanobacteria: The crucial contribution of membrane lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol 1996, 47, 541–568. [Google Scholar]
- Hannah, M.A.; Heyer, A.G.; Hincha, D.K. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet 2005, 1, e26. [Google Scholar]
- Lee, B.H.; Henderson, D.A.; Zhu, J.K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 2005, 17, 3155–3175. [Google Scholar]
- Matsui, A.; Ishida, J.; Morosawa, T.; Mochizuki, Y.; Kaminuma, E.; Endo, T.A.; Okamoto, M.; Nambara, E.; Nakajima, M.; Kawashima, M.; et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 2008, 49, 1135–1149. [Google Scholar]
- Zeller, G.; Henz, S.R.; Widmer, C.K.; Sachsenberg, T.; Ratsch, G.; Weigel, D.; Laubinger, S. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 2009, 58, 1068–1082. [Google Scholar]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–406. [Google Scholar]
- Medina, J.; Bargues, M.; Terol, J.; Perez-Alonso, M.; Salinas, J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 1999, 119, 463–470. [Google Scholar]
- Baker, S.S.; Wilhelm, K.S.; Thomashow, M.F. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol 1994, 24, 701–713. [Google Scholar]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar]
- Canella, D.; Gilmour, S.J.; Kuhn, L.A.; Thomashow, M.F. DNA binding by the Arabidopsis CBF1 transcription factor requires the PKKP/RAGRxKFxETRHP signature sequence. Biochim. Biophys. Acta 2010, 1799, 454–462. [Google Scholar]
- Ohme-Takagi, M.; Shinshi, H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar]
- Franklin, K.A.; Whitelam, G.C. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet 2007, 39, 1410–1413. [Google Scholar]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 2003, 17, 1043–1054. [Google Scholar]
- Miura, K.; Jin, J.B.; Lee, J.; Yoo, C.Y.; Stirm, V.; Miura, T.; Ashworth, E.N.; Bressan, R.A.; Yun, D.J.; Hasegawa, P.M. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 2007, 19, 1403–1414. [Google Scholar]
- Lee, H.; Xiong, L.; Gong, Z.; Ishitani, M.; Stevenson, B.; Zhu, J.K. The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 2001, 15, 912–924. [Google Scholar]
- Ishitani, M.; Xiong, L.; Lee, H.; Stevenson, B.; Zhu, J.K. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 1998, 10, 1151–1161. [Google Scholar]
- Novillo, F.; Alonso, J.M.; Ecker, J.R.; Salinas, J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 3985–3990. [Google Scholar]
- Lazaro, A.; Valverde, F.; Pineiro, M.; Jarillo, J.A. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 2012, 24, 982–999. [Google Scholar]
- Nakamichi, N.; Kusano, M.; Fukushima, A.; Kita, M.; Ito, S.; Yamashino, T.; Saito, K.; Sakakibara, H.; Mizuno, T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 2009, 50, 447–462. [Google Scholar]
- Seo, P.J.; Park, M.J.; Lim, M.H.; Kim, S.G.; Lee, M.; Baldwin, I.T.; Park, C.M. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 2012, 24, 2427–2442. [Google Scholar]
- Cao, S.; Ye, M.; Jiang, S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep 2005, 24, 683–690. [Google Scholar]
- Kendrick, R.E.; Kronenberg, G.H.M. Photomorphogenesis in Plants, 2nd ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994. [Google Scholar]
- Christie, J.M. Phototropin blue-light receptors. Annu. Rev. Plant Biol 2007, 58, 21–45. [Google Scholar]
- Gyula, P.; Schafer, E.; Nagy, F. Light perception and signalling in higher plants. Curr. Opin. Plant Biol 2003, 6, 446–452. [Google Scholar]
- Lin, C.; Shalitin, D. Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol 2003, 54, 469–496. [Google Scholar]
- Franklin, K.A.; Praekelt, U.; Stoddart, W.M.; Billingham, O.E.; Halliday, K.J.; Whitelam, G.C. Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 2003, 131, 1340–1346. [Google Scholar]
- Williams, B.J.; Pellett, N.E.; Klein, R.M. Phytochrome control of growth cessation and initiation of cold acclimation in selected woody plants. Plant Physiol 1972, 50, 262–265. [Google Scholar]
- McKenzie, J.S.; Weiser, C.J.; Burke, M.J. Effects of red and far red light on the initiation of cold acclimation in cornus stolonifera michx. Plant Physiol 1974, 53, 783–789. [Google Scholar]
- Gray, G.R.; Chauvin, L.P.; Sarhan, F.; Huner, N. Cold acclimation and freezing tolerance (A complex interaction of light and temperature). Plant Physiol 1997, 114, 467–474. [Google Scholar]
- Huner, N.P.A.; Quist, G.; Sarhan, F. Energy balance and acclimation to light and cold. Trends Plant Sci 1998, 3, 224–230. [Google Scholar]
- Janda, T.; Szalai, G.; Lesko, K.; Yordanova, R.; Apostol, S.; Popova, L.P. Factors contributing to enhanced freezing tolerance in wheat during frost hardening in the light. Phytochemistry 2007, 68, 1674–1682. [Google Scholar]
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000, 405, 462–466. [Google Scholar]
- Lau, O.S.; Deng, X.W. Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol 2010, 13, 571–577. [Google Scholar]
- Oravecz, A.; Baumann, A.; Mate, Z.; Brzezinska, A.; Molinier, J.; Oakeley, E.J.; Adam, E.; Schafer, E.; Nagy, F.; Ulm, R. Constitutively photomorphogenic1 is required for the UV-B response in Arabidopsis. Plant Cell 2006, 18, 1975–1990. [Google Scholar]
- Yadav, V.; Kundu, S.; Chattopadhyay, D.; Negi, P.; Wei, N.; Deng, X.W.; Chattopadhyay, S. Light regulated modulation of Z-box containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsis. Plant J 2002, 31, 741–753. [Google Scholar]
- Shin, J.; Park, E.; Choi, G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 2007, 49, 981–994. [Google Scholar]
- Harvaux, M.; Kloppstech, K. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta 2001, 213, 953–966. [Google Scholar]
- Wijnen, H.; Young, M.W. Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet 2006, 40, 409–448. [Google Scholar]
- Pruneda-Paz, J.L.; Kay, S.A. An expanding universe of circadian networks in higher plants. Trends Plant Sci 2010, 15, 259–265. [Google Scholar]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 2008, 9, R130. [Google Scholar]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kevei, E.; Toth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar]
- Green, R.M.; Tingay, S.; Wang, Z.Y.; Tobin, E.M. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol 2002, 129, 576–584. [Google Scholar]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000, 290, 2110–2113. [Google Scholar]
- Harmer, S.L. The circadian system in higher plants. Annu. Rev. Plant Biol 2009, 60, 357–377. [Google Scholar]
- Imaizumi, T. Arabidopsis circadian clock and photoperiodism: Time to think about location. Curr. Opin. Plant Biol 2010, 13, 83–89. [Google Scholar]
- Alabadi, D.; Yanovsky, M.J.; Mas, P.; Harmer, S.L.; Kay, S.A. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol 2002, 12, 757–761. [Google Scholar]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.R.; Carre, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2002, 2, 629–641. [Google Scholar]
- Schaffer, R.; Ramsay, N.; Samach, A.; Corden, S.; Putterill, J.; Carre, I.A.; Coupland, G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 1998, 93, 1219–1229. [Google Scholar]
- Wang, Z.Y.; Tobin, E.M. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 1998, 93, 1207–1217. [Google Scholar]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol 2000, 41, 1002–1012. [Google Scholar]
- Alabadi, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Mas, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar]
- Pruneda-Paz, J.L.; Breton, G.; Para, A.; Kay, S.A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 2009, 323, 1481–1485. [Google Scholar]
- Farre, E.M.; Harmer, S.L.; Harmon, F.G.; Yanovsky, M.J.; Kay, S.A. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol 2005, 15, 47–54. [Google Scholar]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar]
- Wang, X.; Ma, L. Unraveling the circadian clock in Arabidopsis. Plant Signal. Behav 2013, 8, e23014. [Google Scholar]
- McClung, C.R. Comes a time. Curr. Opin. Plant Biol 2008, 11, 514–520. [Google Scholar]
- Salome, P.A.; McClung, C.R. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 2005, 17, 791–803. [Google Scholar]
- Bieniawska, Z.; Espinoza, C.; Schlereth, A.; Sulpice, R.; Hincha, D.K.; Hannah, M.A. Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol 2008, 147, 263–279. [Google Scholar]
- Kidokoro, S.; Maruyama, K.; Nakashima, K.; Imura, Y.; Narusaka, Y.; Shinwari, Z.K.; Osakabe, Y.; Fujita, Y.; Mizoi, J.; Shinozaki, K.; et al. The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol 2009, 151, 2046–2057. [Google Scholar]
- Ibanez, C.; Ramos, A.; Acebo, P.; Contreras, A.; Casado, R.; Allona, I.; Aragoncillo, C. Overall alteration of circadian clock gene expression in the chestnut cold response. PLoS One 2008, 3, e3567. [Google Scholar]
- Ramos, A.; Perez-Solis, E.; Ibanez, C.; Casado, R.; Collada, C.; Gomez, L.; Aragoncillo, C.; Allona, I. Winter disruption of the circadian clock in chestnut. Proc. Natl. Acad. Sci. USA 2005, 102, 7037–7042. [Google Scholar]
- James, A.B.; Syed, N.H.; Bordage, S.; Marshall, J.; Nimmo, G.A.; Jenkins, G.I.; Herzyk, P.; Brown, J.W.; Nimmo, H.G. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 2012, 24, 961–981. [Google Scholar]
- Fuchigami, L.H.; Weiser, C.J.; Evert, D.R. Induction of Cold Acclimation in Cornus stolonifera Michx. Plant Physiol 1971, 47, 98–103. [Google Scholar]
- Hellergren, J. Cold Acclimation of suspension cultures of Pinus sylvestris in response to light and temperature treatments. Plant Physiol 1983, 72, 992–995. [Google Scholar]
- Jackson, S.D. Plant responses to photoperiod. New Phytol 2009, 181, 517–531. [Google Scholar]
- Li, C.; Puhakainen, T.; Welling, A.; Viherä-Aarnio, A.; Ernstsen, A.; Junttila, O.; Heino, P.; Palva, E.T. Cold acclimation in silver birch (Betula pendula). Development of freezing tolerance in different tissues and climatic ecotypes. Physiol. Plant 2002, 116, 478–488. [Google Scholar]
- Alonso-Blanco, C.; Gomez-Mena, C.; Llorente, F.; Koornneef, M.; Salinas, J.; Martinez-Zapater, J.M. Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol 2005, 139, 1304–1312. [Google Scholar]
- Mahfoozi, S.; Limin, A.E.; Hayes, P.M.; Hucl, P.; Fowler, D.B. Influence of photoperiod response on the expression of cold hardiness in wheat and barley. Can. J. Plant Sci 2000, 80, 721–724. [Google Scholar]
- Welling, A.; Moritz, T.; Palva, E.T.; Junttila, O. Independent activation of cold acclimation by low temperature and short photoperiod in hybrid aspen. Plant Physiol 2002, 129, 1633–1641. [Google Scholar]
- Heide, O.M.; Prestrud, A.K. Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiol 2005, 25, 109–114. [Google Scholar]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genomics 2011, 12, 30–43. [Google Scholar]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol 2001, 127, 910–917. [Google Scholar]
- Nakashima, K.; Yamaguchi-Shinozaki, K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol. Plant 2006, 126, 62–71. [Google Scholar]
- Hsieh, T.H.; Lee, J.T.; Yang, P.T.; Chiu, L.H.; Charng, Y.Y.; Wang, Y.C.; Chan, M.T. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol 2002, 129, 1086–1094. [Google Scholar]
- Morran, S.; Eini, O.; Pyvovarenko, T.; Parent, B.; Singh, R.; Ismagul, A.; Eliby, S.; Shirley, N.; Langridge, P.; Lopato, S. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J 2011, 9, 230–249. [Google Scholar]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 2003, 33, 751–763. [Google Scholar]
- Oh, S.J.; Kwon, C.W.; Choi, D.W.; Song, S.I.; Kim, J.K. Expression of barley HvCBF4 enhances tolerance to abiotic stress in transgenic rice. Plant Biotechnol. J 2007, 5, 646–656. [Google Scholar]
- Gilmour, S.J.; Sebolt, A.M.; Salazar, M.P.; Everard, J.D.; Thomashow, M.F. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 2000, 124, 1854–1865. [Google Scholar]
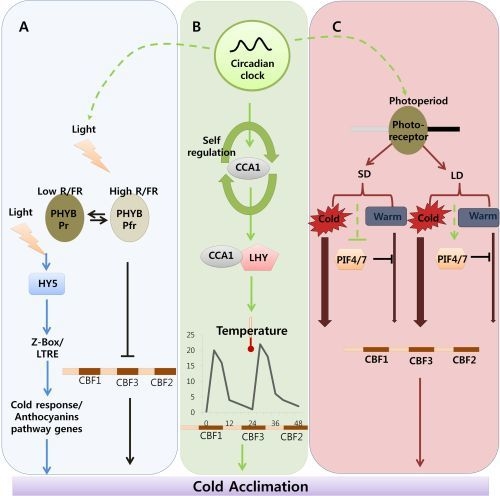
 Indicates cold acclimation pathway in which HY5 regulates the expression of anthocyanin pathway genes through by Z-box/LTRE to control photo-oxidative stress generated by low temperature. → Indicates cold acclimation pathway in which phytochromes negatively regulates the expression of CBF regulon under high red/far red light condition.
Indicates cold acclimation pathway in which HY5 regulates the expression of anthocyanin pathway genes through by Z-box/LTRE to control photo-oxidative stress generated by low temperature. → Indicates cold acclimation pathway in which phytochromes negatively regulates the expression of CBF regulon under high red/far red light condition.
 Indicates circadian clock-regulated cold acclimation pathway in which CCA1 showed selfregulation activity through alternate splicing event in response to cold and it can form heterodimers with LHY to bind directly to promoter region of CBF genes.
Indicates circadian clock-regulated cold acclimation pathway in which CCA1 showed selfregulation activity through alternate splicing event in response to cold and it can form heterodimers with LHY to bind directly to promoter region of CBF genes.
 Indicates influence of circadian clock over light and photoperiod mediated cold acclimation pathway which is unclear.
Indicates influence of circadian clock over light and photoperiod mediated cold acclimation pathway which is unclear.
 Indicates photoperiod involvement in cold acclimation pathway which is regulated by the PHY B and the activity of PIF4/7 in response to SD and LD which can repress the CBF expression under LD by direct binding to promoter region.
Indicates photoperiod involvement in cold acclimation pathway which is regulated by the PHY B and the activity of PIF4/7 in response to SD and LD which can repress the CBF expression under LD by direct binding to promoter region.
 /
/
 Width of arrow indicates the expression levels of CBF genes in response to photoperiod. (For detailed explanation please see the text).
Width of arrow indicates the expression levels of CBF genes in response to photoperiod. (For detailed explanation please see the text).
 Indicates cold acclimation pathway in which HY5 regulates the expression of anthocyanin pathway genes through by Z-box/LTRE to control photo-oxidative stress generated by low temperature. → Indicates cold acclimation pathway in which phytochromes negatively regulates the expression of CBF regulon under high red/far red light condition.
Indicates cold acclimation pathway in which HY5 regulates the expression of anthocyanin pathway genes through by Z-box/LTRE to control photo-oxidative stress generated by low temperature. → Indicates cold acclimation pathway in which phytochromes negatively regulates the expression of CBF regulon under high red/far red light condition.
 Indicates circadian clock-regulated cold acclimation pathway in which CCA1 showed selfregulation activity through alternate splicing event in response to cold and it can form heterodimers with LHY to bind directly to promoter region of CBF genes.
Indicates circadian clock-regulated cold acclimation pathway in which CCA1 showed selfregulation activity through alternate splicing event in response to cold and it can form heterodimers with LHY to bind directly to promoter region of CBF genes.
 Indicates influence of circadian clock over light and photoperiod mediated cold acclimation pathway which is unclear.
Indicates influence of circadian clock over light and photoperiod mediated cold acclimation pathway which is unclear.
 Indicates photoperiod involvement in cold acclimation pathway which is regulated by the PHY B and the activity of PIF4/7 in response to SD and LD which can repress the CBF expression under LD by direct binding to promoter region.
Indicates photoperiod involvement in cold acclimation pathway which is regulated by the PHY B and the activity of PIF4/7 in response to SD and LD which can repress the CBF expression under LD by direct binding to promoter region.
 /
/
 Width of arrow indicates the expression levels of CBF genes in response to photoperiod. (For detailed explanation please see the text).
Width of arrow indicates the expression levels of CBF genes in response to photoperiod. (For detailed explanation please see the text).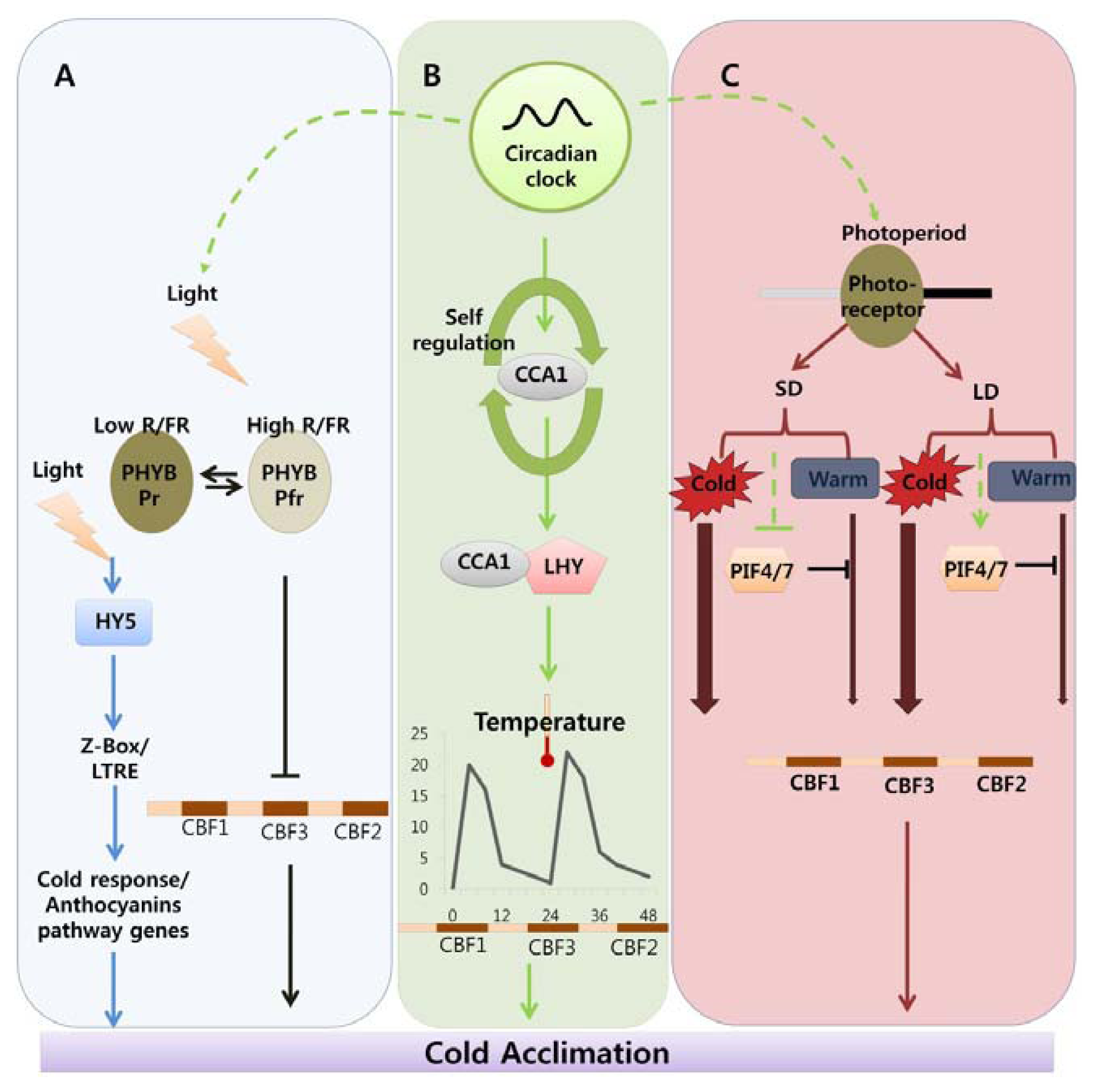
| Symbol | AGI code | Full name | Mechanism and phenotype | Ref. |
|---|---|---|---|---|
| A. Genes involved in light signaling and cold acclimation response pathway | ||||
| CBF2 | AT4G25470 | C-REPEAT/DRE BINDING FACTOR 2 | CBF2 repress the expression of CBF1/3 and the cbf2 mutant showed enhanced cold tolerance | [39] |
| CBF1 | AT4G25490 | C-REPEAT/DRE BINDING FACTOR 1 | CBF1/3 are the positive regulators for cold acclimation and RNAi lines are impaired in cold tolerance | [33] |
| CBF3 | AT4G25480 | C-REPEAT BINDING FACTOR 3 | ||
| HY5 | AT5G11260 | ELONGATED HYPOCOTYL 5 | HY5 positively regulates cold induced gene expression through Z-box/LTRE and hy5 mutant is sensitive | [14] |
| PhyB | AT2G18790 | PHYTOCHROME B | repression of the CBF regulon in high R/FR is mediated by PHYB and PHYD and thus, phyB and phyD mutants showed increased cold tolerance | [34] |
| PhyD | AT4G16250 | PHYTOCHROME D | ||
| B. Genes involved in circadian clock and cold acclimation response pathway | ||||
| PRR5/7/9 | AT5G24470/AT5G02810/AT2G46790 | PSEUDO RESPONSE REGULATOR 5/7/9 | acts as a negative regulators for CBF pathway and thus, triple mutant prr5/7/9 showed increased cold tolerance | [41] |
| CCA1 | AT2G46830 | CIRCADIAN CLOCK ASSOCIATED 1 | CCA1 binds to promoter of CBFs and promotes cold acclimation which is self- regulated in response to cold stress by alternative splicing mechanism | [15,42] |
| LHY | AT1G01060 | LATE ELONGATED HYPOCOTYL | LHY is also important along with CCA1 for circadian regulation of CBFs and double mutant cca1-11/lhy-21 is impaired in freezing tolerance | [15] |
| GI | AT1G22770 | GIGANTEA | GI positively regulates freezing tolerance via a CBF-independent pathway and gi-3 mutant is susceptible to freezing due to impaired sugar metabolism | [43] |
| C. Genes involved in photoperiodism and cold acclimation response pathway | ||||
| PIF4 | AT2G43010 | PHYTOCHROME INTERACTING FACTOR 4 | PIF4 and PIF7 function redundantly and binds to G-box present in the promoter of CBF genes to repress its expression under LD conditions | [19] |
| PIF7 | AT5G61270 | PHYTOCHROME INTERACTING FACTOR 7 | ||
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Maibam, P.; Nawkar, G.M.; Park, J.H.; Sahi, V.P.; Lee, S.Y.; Kang, C.H. The Influence of Light Quality, Circadian Rhythm, and Photoperiod on the CBF-Mediated Freezing Tolerance. Int. J. Mol. Sci. 2013, 14, 11527-11543. https://doi.org/10.3390/ijms140611527
Maibam P, Nawkar GM, Park JH, Sahi VP, Lee SY, Kang CH. The Influence of Light Quality, Circadian Rhythm, and Photoperiod on the CBF-Mediated Freezing Tolerance. International Journal of Molecular Sciences. 2013; 14(6):11527-11543. https://doi.org/10.3390/ijms140611527
Chicago/Turabian StyleMaibam, Punyakishore, Ganesh M. Nawkar, Joung Hun Park, Vaidurya Pratap Sahi, Sang Yeol Lee, and Chang Ho Kang. 2013. "The Influence of Light Quality, Circadian Rhythm, and Photoperiod on the CBF-Mediated Freezing Tolerance" International Journal of Molecular Sciences 14, no. 6: 11527-11543. https://doi.org/10.3390/ijms140611527