1. Introduction
The most important prognostic factor in breast cancer is the presence or absence of disseminated tumours in the axillary lymph nodes [
1,
2] and the spread of cancer to the sentinel lymph nodes, whilst not classifying the patient as having M1 disease, is indicative of a poorer prognosis. Hence the assessment of sentinel lymph node biopsies is the standard of care for staging the axilla [
3–
5], and its intra-operative examination offers the advantage of allowing an immediate axillary dissection of a positive node. This avoids the need for a second operation with its associated anxiety and risk to the individual patient, releases theatre time for more operations, cuts down on the demand for surgical beds and thereby, reduces costs to the National Health Service (NHS).
However, conventional intra-operative methods, such as frozen section and touch cytology, are time- and labour intensive and have a wide range of sensitivity, ranging from 59% to 88.2% for frozen sections and from 40% to 58.3% for touch imprint cytology [
6–
8]. This has led to the evaluation of real time quantitative PCR (qPCR)-based methods, which are sensitive, specific, rapid, relatively inexpensive and can be automated, making their use in the intra-operative setting practical. Furthermore, whereas histology only assesses a representative section of the tissue and therefore introduces sampling error, qPCR analysis can potentially be carried out on the whole node for the presence of markers of interest. The GeneSearch BLNA assay (Veridex LLC, WARREN, NJ, USA) is a now discontinued commercial intra-operative qPCR assay that detected the presence of tumour-specific markers cytokeratin-19 (CK-19) and mammaglobin (MGB) in sentinel lymph nodes [
5,
9–
11]. If either one or both of the markers were positive, the node was considered positive and an axillary clearance would be carried out. Porphobilinogen deaminase (PBGD) was used as a positive control for successful RNA extraction. Despite its proven clinical usefulness, preliminary Federal Drug Administration (FDA) approval and use in the UK and Europe, the assay was withdrawn, mainly due to insufficient implementation in the US home market [
12].
Consequently, we set out to develop a replacement assay, termed Metasin, using the same markers, but with freely available commercial reagents and capable of running on any thermocycler. Importantly, publication of primer and probe details will enable biomedical scientists to optimise the assay within their own institutions. The primers and probe sequences, PCR master mixes and run conditions were optimised and the assay was validated to generate an assay that matched the performance of the GeneSearch assay [
5,
9–
11]. This report compares the performance of the new Metasin assay with both conventional histological methods that provide a ‘gold standard’ as well as the GeneSearch assay for the assessment of archived lymph node lysates from a cohort of 154 cases.
2. Results
2.1. Patient Demographics
Details listing the patient demographics and relevant clinical information were retrospectively collected and are presented in
Table 1. No patient demographics were available for the samples from the Jules Bordet Institute (JBI).
2.2. Characteristics of the PCR Assay
Metasin PCR efficiencies for CK19, MGB and PBGD-specific were 97.3%, 100% and 100%, respectively (results not shown). A comparison of amplification reactions carried out as either monoplex and multiplex reactions resulted in an average difference in quantification cycle (ΔCq) of 1.5, suggesting little interference between the nine primer/probe combinations. Analysis of PCR products on agarose gels resulted in single bands of the expected size and the absence of smearing, indicating an optimised PCR reaction (
Figure S1). Since pseudogenes are known to exist for CK19, its sequence was aligned with those of the pseudogenes, demonstrating the specificity of the primer and probe used (
Figure S2). Absence of pseudogene amplification was also demonstrated empirically by using DNA from 100 colon tumours as templates in the Metasin PCR reaction. No amplification signals were detected in either the CK19 or MGB reactions and the fluorescence detected in the PBGD reaction was below the cut-off threshold established previously. Gel electrophoresis of the PCR reactions showed a series of smears (results not shown).
2.8. Statistical Analysis of JBI Samples
JBI samples were analysed using the same statistical methods, comparing GeneSearch and Metasin Cq values for positive and negative node groups for CK19, MGB and PBGD.
Using the Spearman’s correlation test, there was significant correlation within the positive group between GeneSearch and Metasin for CK19 (rho = 0.842, p < 0.0001) and MGB (rho = 0.454, p < 0.0001) but no significant correlation for PBGD (rho = 0.177, p = 0.101). In the negative group, there was significant correlation for PBGD (rho = 0.383, p < 0.0001) and MGB (rho = 0.804, p < 0.0001), but there was no significant correlation for CK19 (rho = 0, p = 0.997).
2.9. CK19 and MGB IHC
All needle cores of the primary tumours analysed in the retrospective cases at Harlow (n = 154) were stained for CK19 and MGB and evaluated for staining characteristics. These cores were scored positive or negative. Consensus was universal except in 3 cases where there was arbitration by a third pathologist. Two cases failed to show tumours in the material submitted for immunostaining. Of the remaining cases (n = 152), 151 showed CK19 immunoreactivity. Conversely, only 86 of 152 cores (57%) showed MGB immunoreactivity. In two cases where the core biopsies showed intense immuno-reactivity for CK19, RNA from the matched positive lymph node failed to amplify for CK19 but Cq values for MGB were in the range characteristic for micrometastases.
3. Discussion
We describe the initial evaluation of Metasin in parallel with GeneSearch and compared with the benchmark histological assessment. The Metasin assay has many similarities to the withdrawn GeneSearch test, with a similar RNA preparation protocol and choice of predictive markers. It is fast, inexpensive, but can use any qPCR platform, unlike GeneSearch testing, which is software-limited to the Cepheid Smartcycler. Furthermore, in contrast to the commercial assay, we are reporting the primer and probe sequences, so allowing interested laboratories to reproduce and utilise this new assay. The assay’s high sensitivity, specificity and most importantly, capacity to deliver a result in less than 32 min makes it fit for the purpose of application in the intra-operative setting. Furthermore, since results appear in real-time, positive results can be reported as soon as the characteristic amplification plots appear.
Metasin makes use of the same strategy as the GeneSearch assay by including two markers for the prediction of metastatic disease, CK19 and MGB and reporting a positive result if both or either of the two markers generate amplification products. Published data [
16,
17] suggest that 98% to 99% of breast tumours are CK19 positive and extrapolate that the lymph node metastasis from these cases are also likely to be CK19 positive. However, no published data exist that correlate the immunohistochemistry data in the core biopsy or lymph node with the expression of CK19 at the RNA level. Hence it is interesting that during the development of the assay we scored 10% of cases with lymph node metastasis as CK19 negative at the RNA level. Fortunately, these cases were detected by both molecular tests since they were all positive for MGB. Two of these cases showed intense CK19 positivity by immunostaining of the core biopsies. These observations challenge the supposition that it is important to assess CK19 immunostaining of core biopsies for sentinel lymph node analysis [
16,
17]. Clearly, the use of molecular tests targeting CK19 only carries the clinical risk of missing nodal metastasis.
Assays targeting the CK19 gene can result in false positive results due to inadvertent amplification of CK19 pseudogenes, genomic DNA sequences similar to CK19 but not specifying translation products [
18]. Traces of DNA that remain in RNA preparations can serve as templates for amplification with insufficiently validated primers, which are then detected with probes that are not sufficiently specific. Our approach essentially eliminates the risk of pseudogene amplification at two levels: (i) CK19 primer alignment shows several mismatches with both the forward and reverse primer binding sequences, making successful amplification of pseudogenes at the optimised annealing temperatures highly unlikely (see supplementary Figure provided); (ii) the detection probe also has a number of mismatches with pseudogene target sequences. Hence, even if a pseudogene were to be inadvertently amplified, the misamplified PCR amplicons would not be reported at the fluorescence detection temperature.
This was confirmed empirically in three ways:
amplification products run on agarose gels showed only a single band of the correct size (128 bp).
amplification of 100 DNA samples with the Metasin assay did not result in any detectable fluorescence signals and gel electrophoresis of amplified material yielded a series of smears.
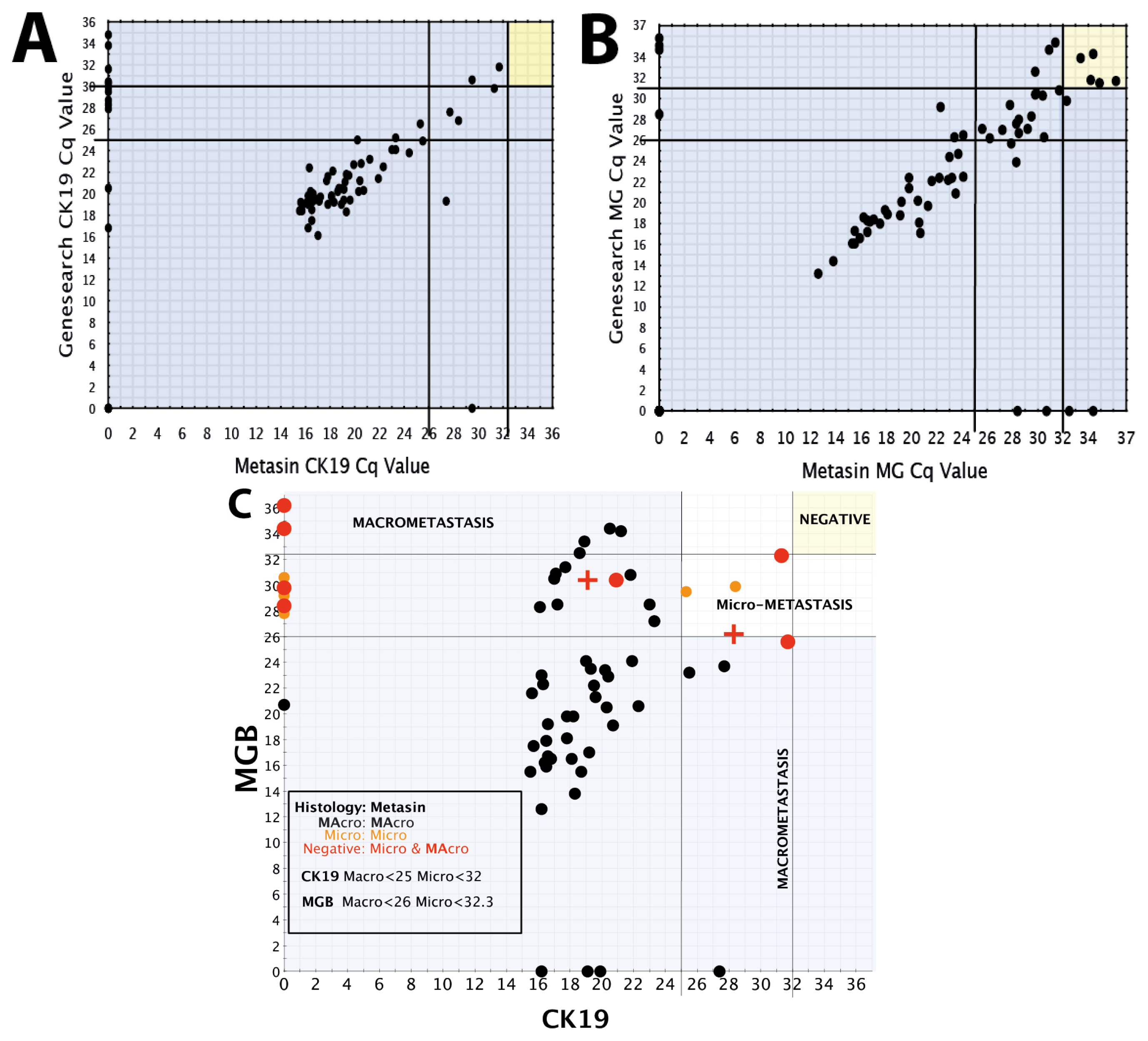
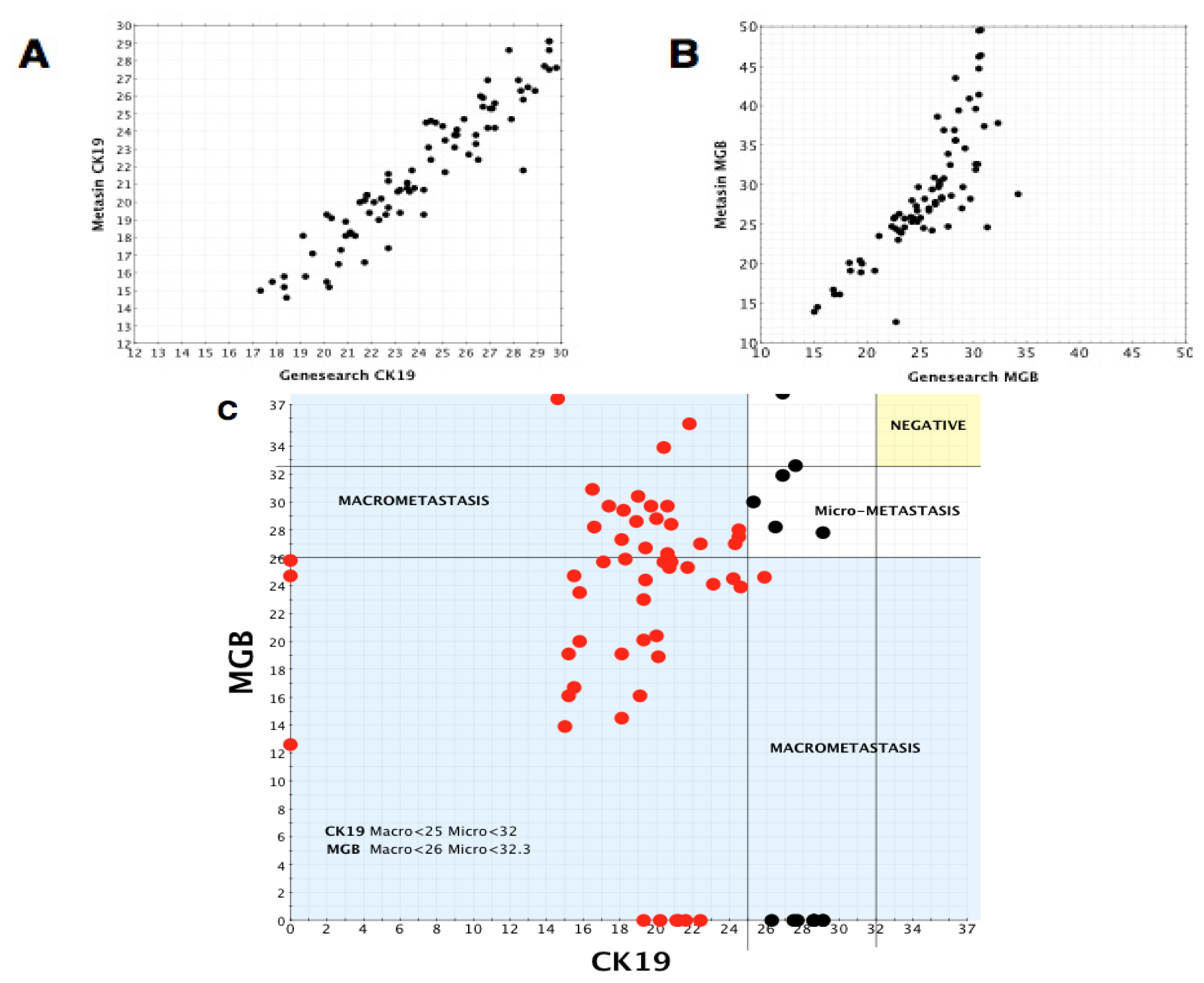
We draw on our observations relating to the low discordant rates of the Metasin assay having analysed over 1700 cases maintaining an overall discordant rate under 4 percent.
Benchmarking the Metasin against the GeneSearch assay clearly demonstrates equivalence of results in most cases. Discrepancies of micrometastasis detection are probably accounted for by variable RNA degradation of the samples obtained from the JBI, which had been stored at −80 °C for more than two years and were used to validate the Metasin assay independently and blinded against the GeneSearch test. Our data reveal a discordance level of 3.98% between Metasin and histology, with a similar level (4.5%) of discordance observed for GeneSearch compared to histology. This is lower than previously reported discordance rates of intraoperative testing using qRT-PCR of up to 7% [
5,
9–
11]. We have recently completed the validation of this assay in a much larger cohort of cases and demonstrate a discordance rate comparable to that reported here of <4% and the assay is now CE marked and available for general use (manuscript in preparation).
Although in diagnostic terms histopathology continues to be the gold standard, it has its limitations. A routine histopathological analysis usually analyses only a 3 μM slice from a wedge of tissue 2000 μM thick; the rest remains unexamined in the paraffin block. Consequently, an examination of sentinel nodes will detect metastases only if tumour is present in that particular 3 μM section. Furthermore, some histological subtypes (lobular carcinoma) are refractory to detection by the naked eye, leaving diagnosis dependent on detection by immuno-staining for cytokeratin [
19]; hence guidelines for breast-cancer-reporting now mandate the use of immunohistochemistry [
20].
To be absolutely certain of the absence of tumour in a lymph node, over 600 3 μM H & E stained sections would need to be carried out through a typical 2 mm lymph node slice [
21]. This is clearly impractical, hence the compromise of examining the lymph nodes at three levels (three level, best practice) instead of examining one slide (single level analysis). Even then, this provides only a “snap-shot” view and most of the original 2 mm thick tissue will remain unexamined in the histology block, making the “gold standard” histological assessment rather inadequate.
Validation studies need to be bench-marked against current practice, which means single level histopathology of sentinel lymph nodes. Hence, alternative slices of serially sectioned lymph nodes were submitted for histology and for molecular analysis. Although examination of lymph nodes at three levels is adequate for most instances, it is not where “disease” is focal. Hence, given the focal nature of metastatic disease in lymph nodes, the separation of the segments makes some discordance between histology and molecular analysis inevitable. For example, one of the cases illustrated here shows a 2.1 mm macrometastasis that was missed by both Metasin and by GeneSearch. Examination of multiple levels revealed this to be focal disease with a macro-metastasis present in the horizontal plane, which cuts out after the examination of 2 levels (300 μM) and is not present in the third level.
Unlike PCR-based assays for sentinel node assessment, the OSNA assay available for intraoperative testing of sentinel nodes advocates the use of the whole node [
22]. Here, we have demonstrated that the molecular approach can theoretically detect the presence of 500 to 1000 cells in lymph node homogenate without being subject to visual interpretive bias. Hence there is a rational argument for using the whole node for molecular analysis. However, this leaves no reserve material available for review and so requires any molecular assay to be utterly reliable. Metasin failed to detect three macro-metastasis (see Table provided in supplementary data), two of which also tested negative by the Genesearch test, suggesting that this discordance was due to allocation bias (see Table provided in supplementary data) and that the assays would have detected the tumour had the whole node been analysed. One was detected by the Genesearch assay, with the reason for this discrepancy unclear. Assay failure occurs in around 1.5% of cases (manuscript in preparation) and there is a rare theoretical risk of false positives due to the presence of epithelial inclusions [
23,
24] or the expression of CK19 in some high-grade lymphomas [
25,
26]. Clearly, these limitations must be highlighted in guidance notes to clinicians.
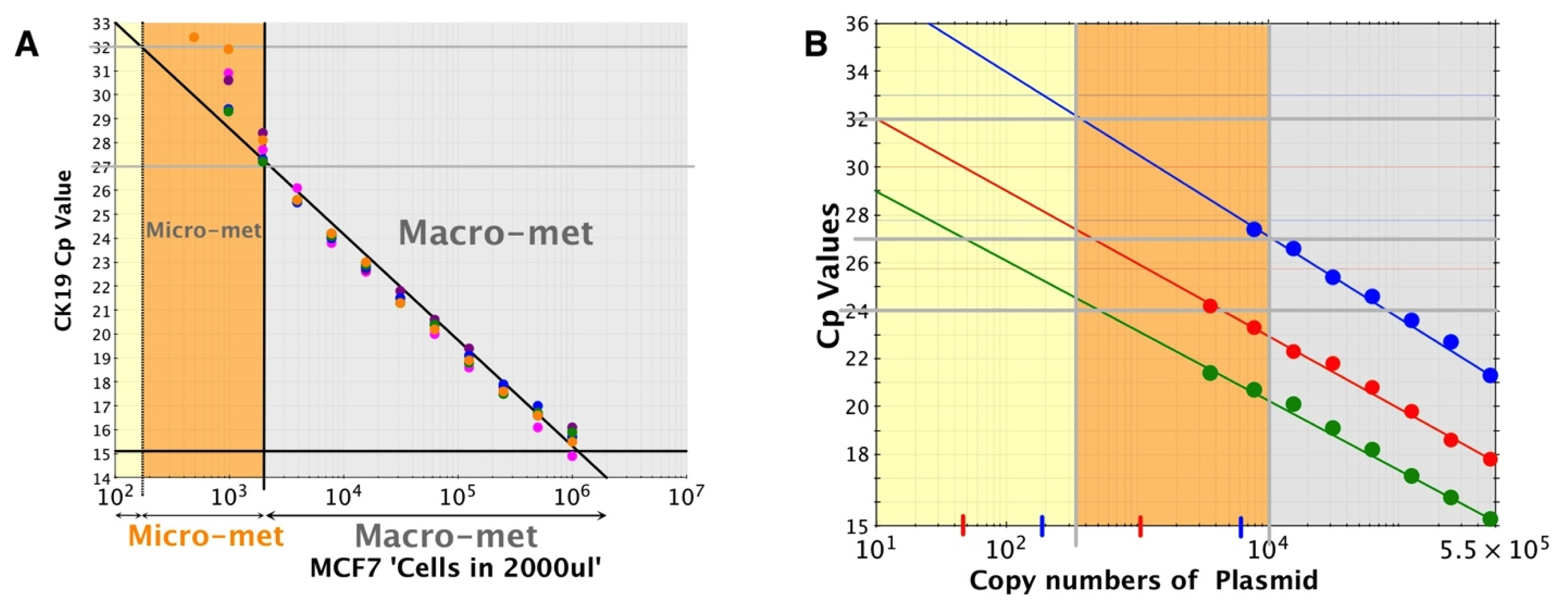
The sensitivity of the assay was determined by using doubling dilutions of cell line RNA or plasmid DNA, since the control MCF7 cell line expresses only CK19 and PBGD, but not MGB. The assay is capable of detecting as few as 200 cell equivalents of RNA, although, this is not consistently reproduced. The significance of these observations is that ITCs are estimated to be of the order of 200 to 500 cells since counts more than this most likely represent micro-metastasis [
5,
27,
28].
4. Experimental Section
4.1. Ethical Approval/PAH Samples
This study was ethically approved by the Essex 2 Research Ethics Committee (Ethics Approval Reference: 07/H0302/129 & REC 10/H301/24). All patients undergoing sentinel lymph node biopsy for breast cancer during March 2008 to July 2010 at PAH were consented for their sentinel lymph node tissue to be tested using the GeneSearch and Metasin tests. The inclusion criteria for participants included those undergoing a surgical excision of a primary operable breast cancer and sentinel lymph node sampling. No exclusion criteria were named. The patients were operated on by surgeons after the implementation of the “NEW START” program [
4]. The results of primary data relating to the GeneSearch and the histology were known to the operators before validating the assay.
4.2. Histopathological Processing and Assessment of Sentinel Lymph Nodes
The sentinel lymph nodes were identified by a combined technique of radioactive (TcM99m) labelled nanocolloid and 2 mL of diluted Patent V blue dye [
29]. Excess fat was removed from the lymph nodes and they were sliced into 2 mm sections with alternate slices allocated to Genesearch/Metasin and histology assessment. Slices for histology were processed individually according to local histology protocol and involved cutting five levels of 100 μM each together with three unstained serial sections per level. The slides were examined by two experienced Consultant Histopathologists and discordant node analysis included examination of a further five levels with immunostaining for MNF116 [
30].
4.4. Ethical Approval/JBI RNA
NREC project REC 10/H301/24 covered ethical approval for RNA obtained from the JBI. Stored frozen (−80 °C) RNA from 193 nodes were obtained retrospectively. These had been analysed previously using the GeneSearch assay, which recorded 104 nodes as negative and 89 as positive. The demographic details, histological outcome and axillary clearance data for this grouping were incomplete. The operators were blinded to the results of the original GeneSearch assay, which were provided after conclusion of the analysis.
4.5. Quantitation and Identification of Template (RNA and DNA)
A Nanodrop microvolume spectrophotometer (Thermo Scientific, Wilmington, DE, USA) was used to quantify all RNA samples. Purity was assessed using 260/280 ratios and, in addition, the RNA quality of a selection of random samples was assessed using a Bioanalyzer 2100 (Agilent, Stockport, UK).
4.6. Primer and Probe Sequences
Target sequences for PBGD (NM_000190), CK19 (NM_002276) and MGB (NM_002411) were obtained from the National Centre for Biotechnology Information (NCBI) GenBank sequence database (
http://www.ncbi.nlm.nih.gov/genbank/). qPCR amplification primers and hydrolysis probes were designed by a specialist oligonucleotide supply company (TIB Molbiol, Berlin, Germany) (
Table 7). All primers were located in different exons, thus minimising the risk of genomic DNA amplification and great care was taken to ensure that pseudogenes [
18] are not amplified (supplementary data). Primer annealing temperatures ranged from 56.2 to 57.9 °C.
4.7. RT-PCR Assay
All PCR assays were performed on the SmartCycler 2.0 (Cepheid, Sunnyvale, CA, USA) with the aim of generating quantification cycles (Cqs) for Metasin that are close to those of the Genesearch test. Hence, optimisation matrices of primer (1.0–2.5 μM) and probe (0.05–0.1 μM) concentrations, together with optimised RNA concentrations were used to identify the optimal combination of oligonucleotide and RNA concentrations. The final protocol was a one-tube, two-step protocol that makes use of the dual RNA- and DNA-dependent polymerase activities of Tth DNA polymerase. Reactions used 4 μL RNA in a total reaction volume of 25 μL with final primer and probe concentrations of 2.5 μM and 1.0 μM, respectively and were run in parallel by a Biomedical Scientist (BMS) (PSG) and a research fellow (SAR). The RT step of the two-step protocol was carried out for 4 min at 60 °C, followed by 40 cycles of 95 °C 10 s, target-specific annealing temperature for 10 s and a polymerisation step at 72 °C for 10 s. The details of the reporting of the RT-qPCR experiments described here are compliant with the MIQE guidelines [
32].
4.8. Positive and Negative Controls
Every run contained at least one positive control and one negative control. The positive control consisted of a sample of RNA that had previously tested positive for all three mRNA targets; the negative control was an RNA sample that had previously tested negative for CK19 and MGB, but positive for PBGD.
4.9. PCR Efficiency and Copy Number Calculations
The MCF7 (ATCC) cell line was grown in Eagle’s MEM culture medium supplemented by 10% FBS in 5% CO
2[
33]. On reaching 70–80 percent confluence, cells were trypsinised, harvested and counted. One million cells were used for RNA extractions using the modified RNA extraction protocol described above. RNA was two-fold serially diluted, 4 μL of each serially diluted RNA was used with the Metasin assay and resulting Cqs were plotted against the log of RNA concentration. This was repeated five times and results are shown in
Figure 3A.
To correlate Cq with copy number, linearised plasmids specifying the amplicons for CK19, MGB or PBGD were prepared by TiBMOL BIOL at 4 × 10
9 copies/μL. They were ten-fold serially diluted, amplified using the Metasin assay and resulting Cq values were plotted against the log of the copy numbers (
Figure 3B).
4.10. Cut-Off Cq Value for CK19 and MGB for Positive and Negative Nodes
A scatter plot was prepared plotting Metasin Cq values for CK19 vs. MGB of all nodes. Each point on the scatter plot was also correlated with histology. Vertical and horizontal lines were drawn selecting the best Cq value for both markers which incorporated all the true positive nodes without creating any false-positive node results. Once these cut-offs were established, their sensitivities along with the confidence intervals and specificities were calculated as well as their negative and positive predictive values. The cut off Cq value for each marker was chosen so as to minimise the number of false positive results.
The same method was used to determine the optimum cut-offs for micrometastases and macrometastases.
4.11. Statistical Analysis
The data were shown to be non-parametric, with the Kolmogorov-Smirnoff test of distribution indicating a uniform but non-Gaussian distribution (Z value = 2.187, p value < 0.0001). Cq values were separated according to positive and negative nodes and comparison of Genesearch and Metasin for PBGD, CK19 and MGB was performed using Spearman correlation tests. The null hypothesis was defined as there not being any significant difference between Genesearch and Metasin Cq values for each of the three markers.
4.12. CK19 and MGB IHC on Breast Cores
All needle cores of the cases analysed in the retrospective cases at Harlow were stained for CK19 and MGB evaluated for staining characteristics. These cores were scored positive or negative by two experienced Consultant Histopathologists (EA & MLS). The Consultant Histopathologist reviewing the slides were “blind” to the outcome of the Metasin and GeneSearch data.