The Enzyme-Mediated Direct Reversal of a Dithymine Photoproduct in Germinating Endospores
Abstract
:1. The Unique Photo-Damage in Bacterial Endospores
2. SP Repair by NER and SPL
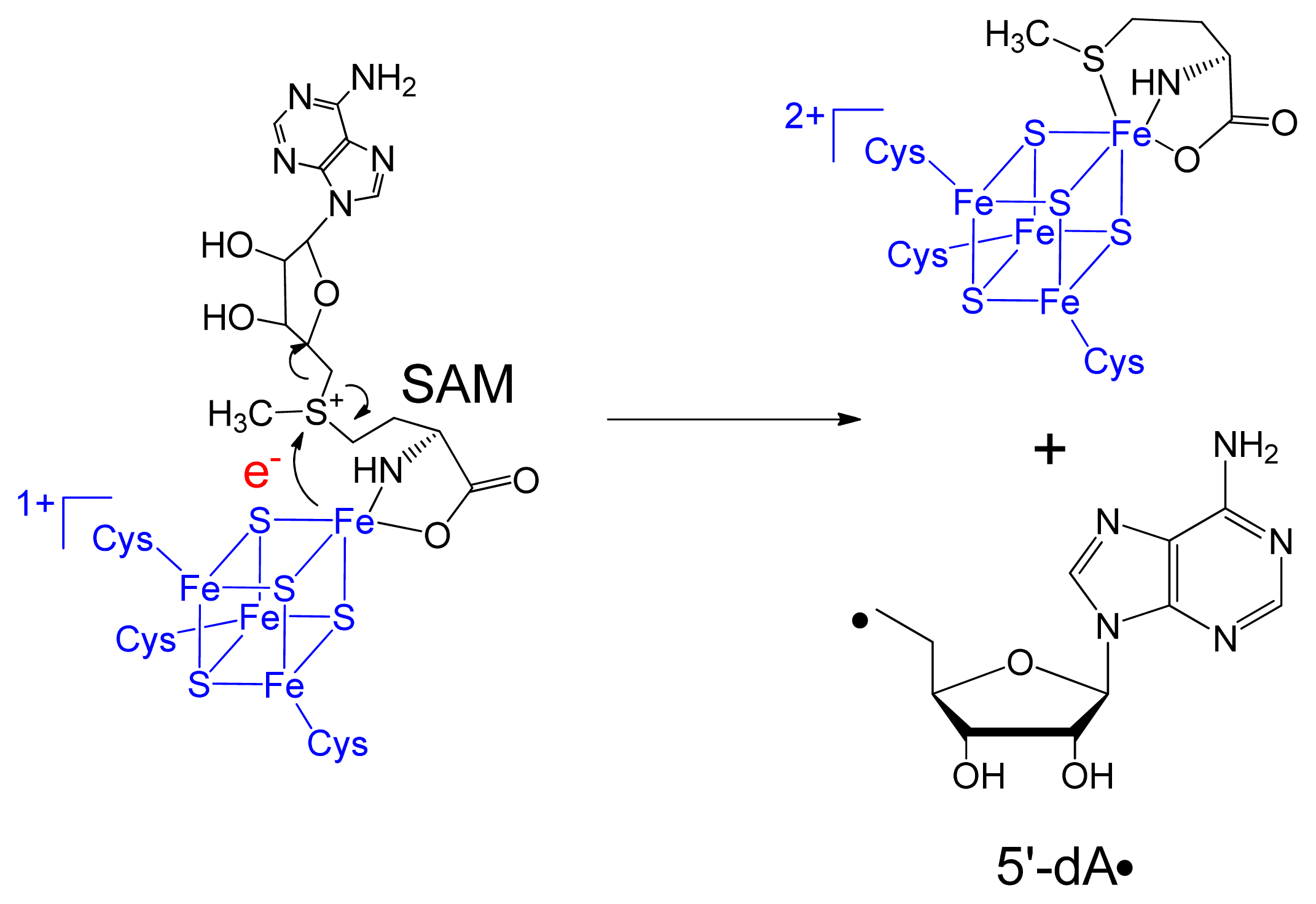
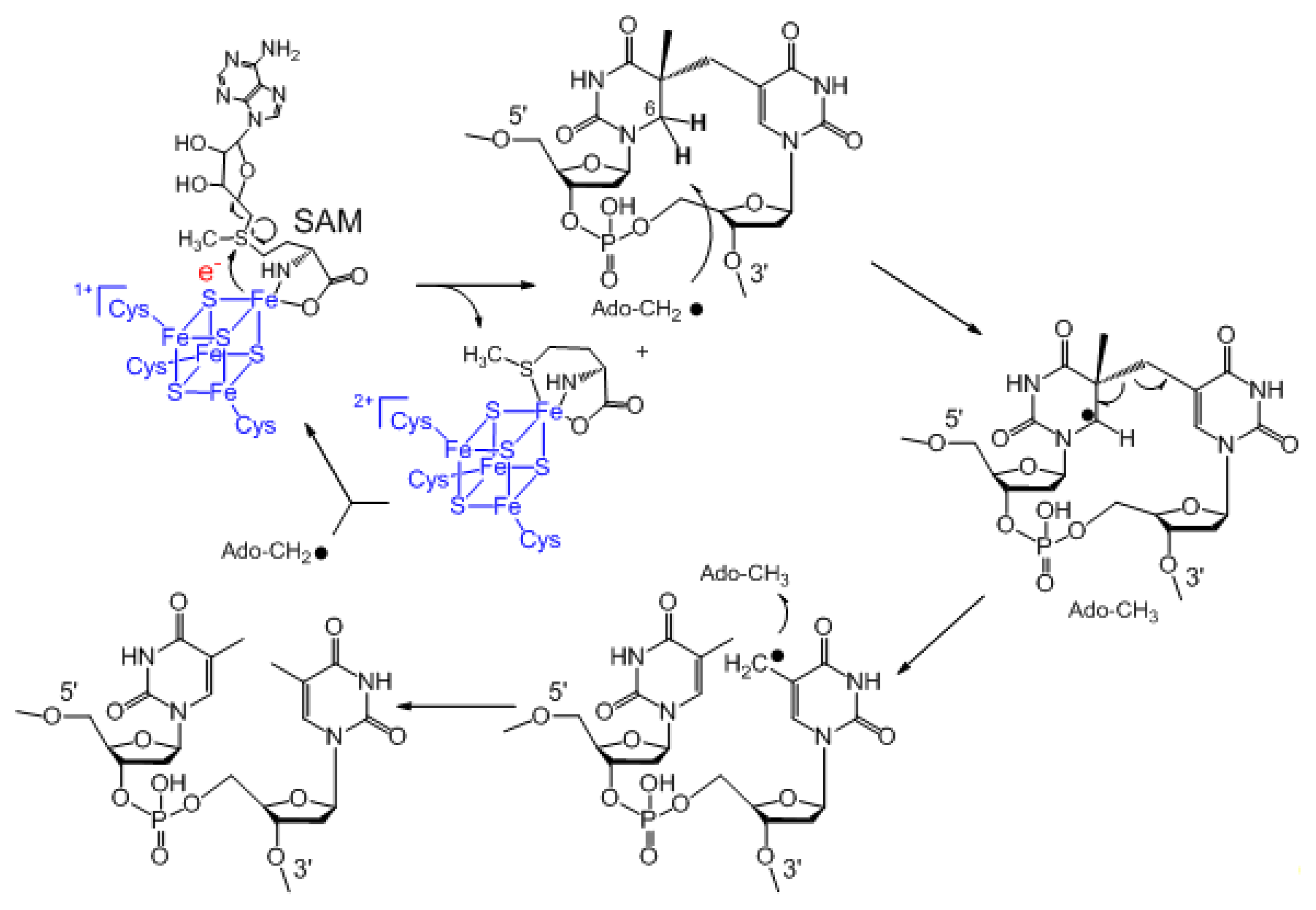
3. Mechanism of SPL Repair
4. The Stereoselectivity of SPL Reaction
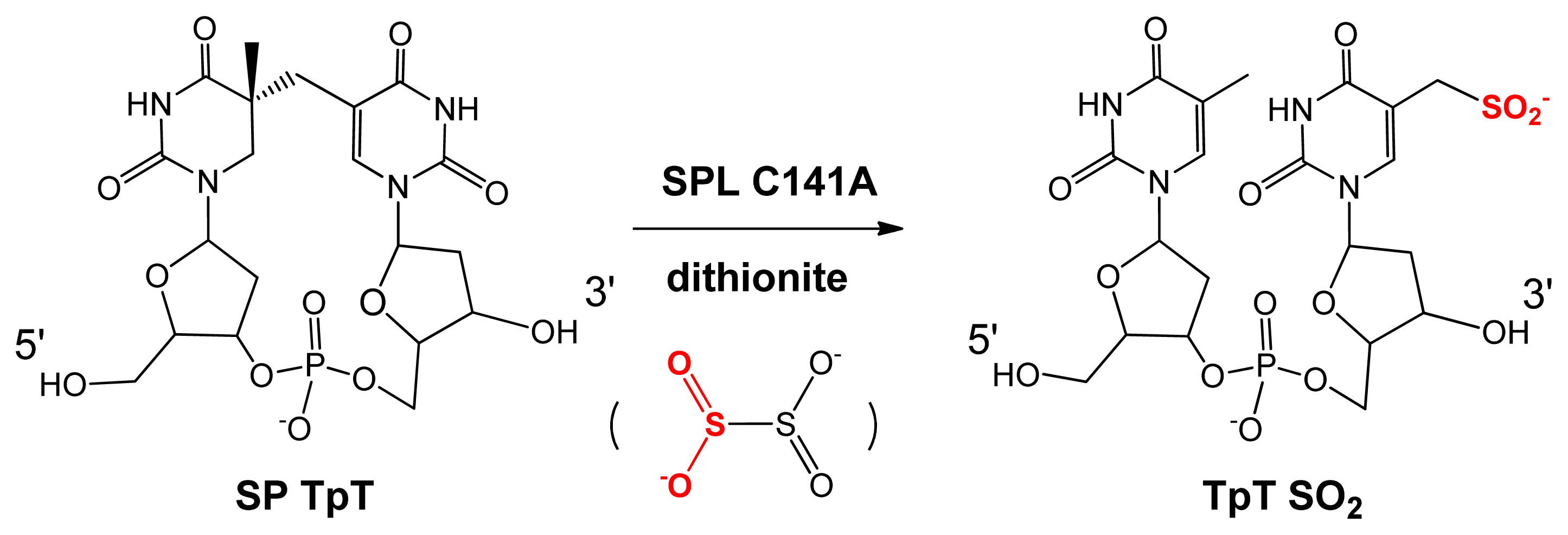
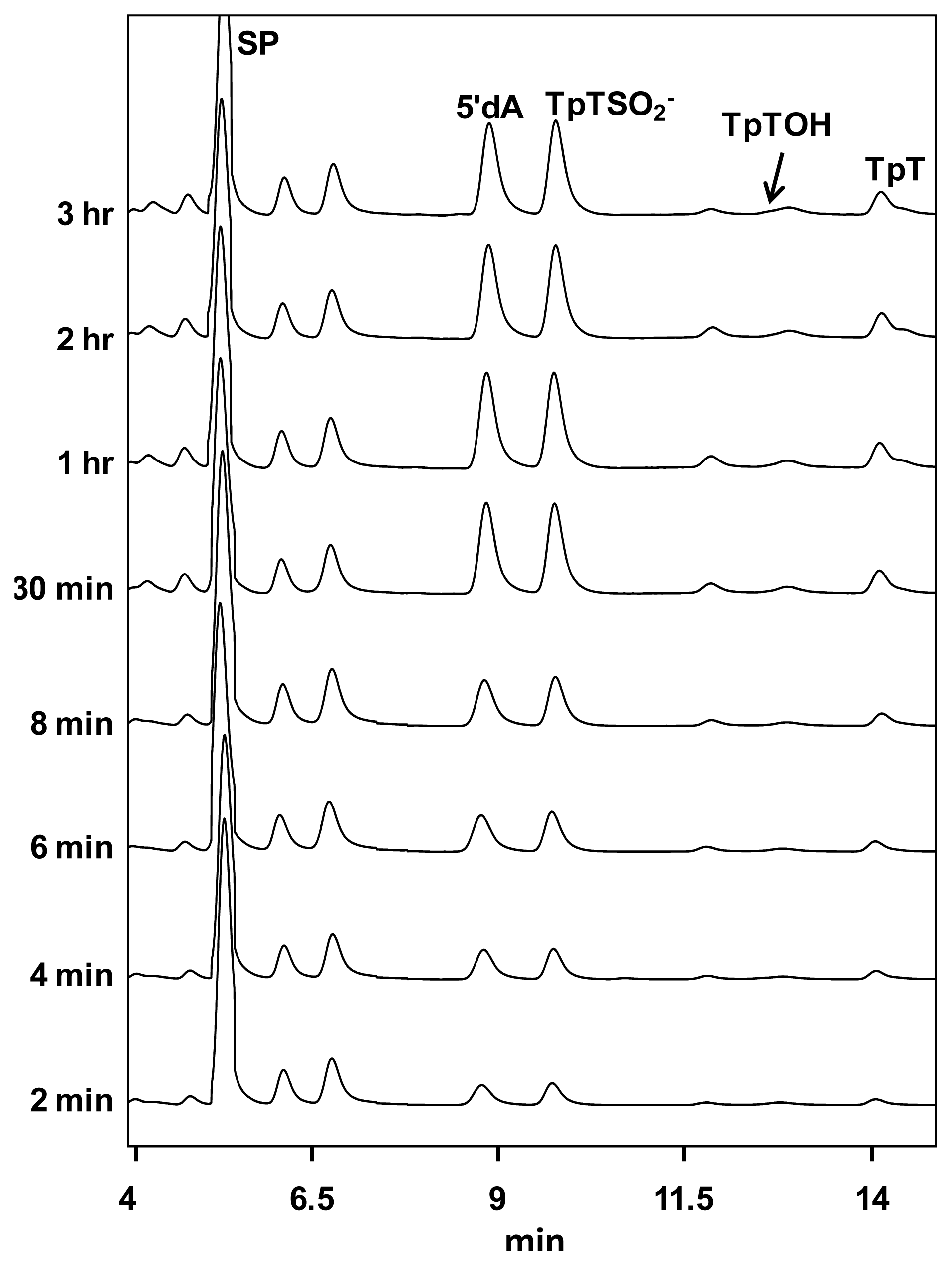
5. SPL Catalysis Requires an Essential Cysteine
6. The Presence of a Novel Radical Transfer Pathway Mediated by Tyrosine Residues in SPL
7. Summary and Outlook
Acknowledgements
Conflict of Interest
References
- Nicholson, W.; Schuerger, A.; Setlow, P. The solar UV environment and bacterial spore UV resistance: considerations for Earth-to-Mars transport by natural processes and human spaceflight. Mutat. Res. Fundam. Mol. Mech. Mutagen 2005, 571, 249–264. [Google Scholar]
- Prescott, L.M.; Harley, J.P.; Klein, D.A. Microbiology, 6th ed; McGraw-Hill Higher Education: Dubuque, IA, USA, 2005. [Google Scholar]
- Lamola, A.A. Triplet photosensitization and the photobiology of thymine dimers in DNA. Pure Appl. Chem 1970, 24, 599–610. [Google Scholar]
- Cadet, J.; Vigny, P. Photochemistry and the Nucleic Acids; Wiley: New York, NY, USA, 1990; Volume 1. [Google Scholar]
- Durbeej, B.; Eriksson, L.A. On the formation of cyclobutane pyrimidine dimers in UV-irradiated DNA: Why are thymines more reactive? Photochem. Photobiol 2003, 78, 159–167. [Google Scholar]
- Lee, K.S.; Bumbaca, D.; Kosman, J.; Setlow, P.; Jedrzejas, M.J. Structure of a protein-DNA complex essential for DNA protection in spores of Bacillus species. Proc. Natl. Acad. Sci. USA 2008, 105, 2806–2811. [Google Scholar]
- Bumbaca, D.; Kosman, J.; Setlow, P.; Henderson, R.K.; Jedrzejas, M.J. Crystallization and preliminary X-ray analysis of the complex between a Bacillus subtilis [alpha]/[beta]-type small acid-soluble spore protein and DNA. Acta Crystallogr. F 2007, 63, 503–506. [Google Scholar]
- Mohr, S.C.; Sokolov, N.V.; He, C.M.; Setlow, P. Binding of small acid-soluble spore proteins from Bacillus subtilis changes the conformation of DNA from B to A. Proc. Natl. Acad. Sci. USA 1991, 88, 77–81. [Google Scholar]
- Setlow, P. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol 1995, 49, 29–54. [Google Scholar]
- Desnous, C.L.; Guillaume, D.; Clivio, P. Spore photoproduct: A key to bacterial eternal life. Chem. Rev 2010, 110, 1213–1232. [Google Scholar]
- Setlow, B.; Setlow, P. Dipicolinic acid greatly enhances production of spore photoproduct in bacterial spores upon UV irradiation. Appl. Environ. Microbiol 1993, 59, 640–643. [Google Scholar]
- Moeller, R.; Douki, T.; Cadet, J.; Stackebrandt, E.; Nicholson, W.L.; Rettberg, P.; Reitz, G.; Horneck, G. UV-radiation-induced formation of DNA bipyrimidine photoproducts in Bacillus subtilis endospores and their repair during germination. Int. Microbiol 2007, 10, 39–46. [Google Scholar]
- Slieman, T.A.; Nicholson, W.L. Artificial and solar UV radiation induces strand breaks and cyclobutane pyrimidine dimers in Bacillus subtilis spore DNA. Appl. Environ. Microbiol 2000, 66, 199–205. [Google Scholar]
- Nicholson, W.L.; Setlow, B.; Setlow, P. Ultraviolet irradiation of DNA complexed with alpha/beta-type small, acid-soluble proteins from spores of Bacillus or Clostridium species makes spore photoproduct but not thymine dimers. Proc. Natl. Acad. Sci. USA 1991, 88, 8288–8292. [Google Scholar]
- Lin, G.; Li, L. Elucidation of spore-photoproduct formation by isotope labeling. Angew. Chem. Int. Ed 2010, 49, 9926–9929. [Google Scholar]
- Donnellan, J.E.; Stafford, R.S. The ultraviolet photochemistry and photobiology of vegetative cells and spores of Bacillus megaterium. Biophys. J 1968, 8, 17–28. [Google Scholar]
- Munakata, N. Mapping of the genes controlling excision repair of Pyrimidine photoproducts in Bacillus subtilis. Mol. Gen. Genet 1977, 156, 49–54. [Google Scholar]
- Fajardo-Cavazos, P.; Salazar, C.; Nicholson, W.L. Molecular cloning and characterization of the Bacillus subtilis spore photoproduct lyase (spl) gene, which is involved in repair of UV radiation-induced DNA damage during spore germination. J. Bacteriol 1993, 175, 1735–1744. [Google Scholar]
- Munakata, N.; Rupert, C.S. Genetically controlled removal of spore photoproduct from deoxyribonucleic acid of ultraviolet-irradiated Bacillus subtilis Spores. J. Bacteriol 1972, 111, 192–198. [Google Scholar]
- Munakata, N.; Rupert, C.S. Dark repair of DNA containing “spore photoproduct” in Bacillus subtilis. Mol. Gen. Genet 1974, 130, 239–250. [Google Scholar]
- Wang, T.-C.V.; Rupert, C.S. Evidence for the monomerization of spore photoproduct to two thymines by the light-independent “spore repair” process in Bacillus subtilis. Photochem. Photobiol 1977, 25, 123–127. [Google Scholar]
- Munakata, N. Genetic analysis of a mutant of Bacillus subtilis producing ultraviolet-sensitive spores. Mol. Gen. Genet 1969, 104, 258–263. [Google Scholar]
- Pedraza-Reyes, M.; Gutierrez-Corona, F.; Nicholson, W.L. Temporal regulation and forespore-specific expression of the spore photoproduct lyase gene by sigma-G RNA polymerase during Bacillus subtilis sporulation. J. Bacteriol 1994, 176, 3983–3991. [Google Scholar]
- Benjdia, A. DNA photolyases and SP lyase: structure and mechanism of light-dependent and independent DNA lyases. Curr. Opin. Struct. Biol 2012, 22, 711–720. [Google Scholar]
- Sancar, A. Structure and Function of DNA Photolyase and Cryptochrome Blue-Light Photoreceptors. Chem. Rev 2003, 103, 2203–2238. [Google Scholar]
- Chandor, A.; Berteau, O.; Douki, T.; Gasparutto, D.; Sanakis, Y.; Ollagnier-De-Choudens, S.; Atta, M.; Fontecave, M. Dinucleotide spore photoproduct, a minimal substrate of the DNA repair spore photoproduct lyase enzyme from Bacillus subtilis. J. Biol. Chem 2006, 281, 26922–26931. [Google Scholar]
- Slieman, T.A.; Rebeil, R.; Nicholson, W.L. Spore photoproduct (SP) lyase from Bacillus subtilis specifically binds to and cleaves SP (5-thyminyl-5,6-dihydrothymine) but not cyclobutane pyrimidine dimers in UV-irradiated DNA. J. Bacteriol 2000, 182, 6412–6417. [Google Scholar]
- Donnellan, J.E., Jr; Setlow, R.B. Thymine photoproducts but not thymine dimers found in ultraviolet-irradiated bacterial spores. Science 1965, 149, 308–310. [Google Scholar]
- Setlow, P. Small, acid-soluble spore proteins of Bacillus species: Structure, synthesis, genetics, function, and degradation. Annu. Rev. Microbiol 1988, 42, 319–338. [Google Scholar]
- Setlow, P. Resistance of spores of Bacillus species to ultraviolet light. Environ. Mol. Mutagen 2001, 38, 97–104. [Google Scholar]
- Nicholson, W.L.; Munakata, N.; Horneck, G.; Melosh, H.J.; Setlow, P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev 2000, 64, 548–572. [Google Scholar]
- Li, L. Mechanistic studies of the radical SAM enzyme spore photoproduct lyase (SPL). Biochim. Biophys. Acta 2012, 1824, 1264–1277. [Google Scholar]
- Sofia, H.J.; Chen, G.; Hetzler, B.G.; Reyes-Spindola, J.F.; Miller, N.E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res 2001, 29, 1097–1106. [Google Scholar]
- Marsh, E.N.G.; Patterson, D.P.; Li, L. Adenosyl radical: Reagent and catalyst in enzyme reactions. Chem. Biol. Chem 2010, 11, 604–621. [Google Scholar]
- Chatterjee, A.; Li, Y.; Zhang, Y.; Grove, T.L.; Lee, M.; Krebs, C.; Booker, S.J.; Begley, T.P.; Ealick, S.E. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat. Chem. Biol 2008, 4, 758–765. [Google Scholar]
- Paraskevopoulou, C.; Fairhurst, S.A.; Lowe, D.J.; Brick, P.; Onesti, S. The elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol. Microbiol 2006, 59, 795–806. [Google Scholar]
- Walsby, C.J.; Ortillo, D.; Yang, J.; Nnyepi, M.R.; Broderick, W.E.; Hoffman, B.M.; Broderick, J.B. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “radical-Sam” protein superfamily. Inorg. Chem 2005, 44, 727–741. [Google Scholar]
- Booker, S.J. Radical SAM enzymes and radical enzymology. Biochim. Biophys. Acta 2012, 1824, 1151–1153. [Google Scholar]
- Frey, P.A.; Hegeman, A.D.; Ruzicka, F.J. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol 2008, 43, 63–88. [Google Scholar]
- Driesener, R.; Challand, M.; Mcglynn, S.; Shepard, E.; Boyd, E.; Broderick, J.; Peters, J.; Roach, P. [FeFe]-Hydrogenase cyanide ligands derived from S-Adenosylmethionine-Dependent cleavage of tyrosine. Angew. Chem. Int. Ed. 2010, 49, 1687–1690. [Google Scholar]
- Mulder, D.W.; Boyd, E.S.; Sarma, R.; Lange, R.K.; Endrizzi, J.A.; Broderick, J.B.; Peters, J.W. Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA[Dgr]EFG. Nature 2010, 465, 248–251. [Google Scholar]
- Cicchillo, R.M.; Iwig, D.F.; Jones, A.D.; Nesbitt, N.M.; Baleanu-Gogonea, C.; Souder, M.G.; Tu, L.; Booker, S.J. Lipoyl synthase requires two equivalents of S-Adenosyl-L-methionine to synthesize one equivalent of lipoic acid. Biochemistry 2004, 43, 6378–6386. [Google Scholar]
- Boal, A.K.; Grove, T.L.; Mclaughlin, M.I.; Yennawar, N.H.; Booker, S.J.; Rosenzweig, A.C. Structural Basis for Methyl Transfer by a Radical SAM Enzyme. Science 2011, 332, 1089–1092. [Google Scholar]
- Grove, T.L.; Benner, J.S.; Radle, M.I.; Ahlum, J.H.; Landgraf, B.J.; Krebs, C.; Booker, S.J. A radically different mechanism for S-Adenosylmethionine–Dependent methyltransferases. Science 2011, 332, 604–607. [Google Scholar]
- Ruszczycky, M.W.; Choi, S.H.; Liu, H.W. EPR-kinetic isotope effect study of the mechanism of radical-mediated dehydrogenation of an alcohol by the radical SAM enzyme DesII. Proc. Natl. Acad. Sci. USA 2013, 110, 2088–2093. [Google Scholar]
- Szu, P.-H.; Ruszczycky, M.W.; Choi, S.-H.; Yan, F.; Liu, H.-W. Characterization and mechanistic studies of DesII: A radical S-Adenosyl-L-methionine enzyme involved in the biosynthesis of TDP-D-Desosamine. J. Am. Chem. Soc 2009, 131, 14030–14042. [Google Scholar]
- Ruszczycky, M.W.; Choi, S.-H.; Liu, H.-W. Stoichiometry of the redox neutral deamination and oxidative dehydrogenation reactions catalyzed by the radical SAM enzyme DesII. J. Am. Chem. Soc 2010, 132, 2359–2369. [Google Scholar]
- Forouhar, F.; Arragain, S.; Atta, M.; Gambarelli, S.; Mouesca, J.-M.; Hussain, M.; Xiao, R.; Kieffer-Jaquinod, S.; Seetharaman, J.; Acton, T.B.; et al. Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. Nat. Chem. Biol 2013, 9, 333–338. [Google Scholar]
- Pierre, S.; Guillot, A.; Benjdia, A.; Sandström, C.; Langella, P.; Berteau, O. Thiostrepton tryptophan methyltransferase expands the chemistry of radical SAM enzymes. Nat. Chem. Biol 2012, 8, 957–959. [Google Scholar]
- Decamps, L.; Philmus, B.; Benjdia, A.; White, R.; Begley, T.P.; Berteau, O. Biosynthesis of F0, precursor of the F420 cofactor, requires a unique two radical-SAM domain enzyme and tyrosine as substrate. J. Am. Chem. Soc 2012, 134, 18173–18176. [Google Scholar]
- Kim, H.J.; Mccarty, R.M.; Ogasawara, Y.; Liu, Y.-N.; Mansoorabadi, S.O.; Levieux, J.; Liu, H.-W. GenK-Catalyzed C-6′ Methylation in the Biosynthesis of Gentamicin: Isolation and Characterization of a Cobalamin-Dependent Radical SAM Enzyme. J. Am. Chem. Soc. 2013. [Google Scholar] [CrossRef]
- Benjdia, A.; Leprince, J.R.M.; Sandström, C.; Vaudry, H.; Berteau, O. Mechanistic investigations of anaerobic sulfatase-maturating enzyme: Direct Cβ H-Atom abstraction catalyzed by a radical AdoMet enzyme. J. Am. Chem. Soc 2009, 131, 8348–8349. [Google Scholar]
- Rebeil, R.; Sun, Y.; Chooback, L.; Pedraza-Reyes, M.; Kinsland, C.; Begley, T.P.; Nicholson, W.L. Spore photoproduct lyase from Bacillus subtilis spores is a novel iron-sulfur DNA repair enzyme which shares features with proteins such as class III anaerobic ribonucleotide reductases and pyruvate-formate lyases. J. Bacteriol 1998, 180, 4879–4885. [Google Scholar]
- Rebeil, R.; Nicholson, W.L. The subunit structure and catalytic mechanism of the Bacillus subtilis DNA repair enzyme spore photoproduct lyase. Proc. Natl. Acad. Sci. USA 2001, 98, 9038–9043. [Google Scholar]
- Mehl, R.A.; Begley, T.P. Mechanistic studies on the repair of a novel DNA photolesion: The spore photoproduct. Org. Lett 1999, 1, 1065–1066. [Google Scholar]
- Cheek, J.; Broderick, J. Direct H atom abstraction from spore photoproduct C-6 initiates DNA repair in the reaction catalyzed by spore photoproduct lyase: Evidence for a reversibly generated adenosyl radical intermediate. J. Am. Chem. Soc 2002, 124, 2860–2861. [Google Scholar]
- Frey, P.; Magnusson, O. S-Adenosylmethionine: A wolf in sheep’s clothing, or a rich man’s adenosylcobalamin? Chem. Rev 2003, 103, 2129–2148. [Google Scholar]
- Chen, D.; Walsby, C.; Hoffman, B.M.; Frey, P.A. Coordination and mechanism of reversible cleavage of S-adenosylmethionine by the [4Fe-4S] center in lysine 2,3-aminomutase. J. Am. Chem. Soc 2003, 125, 11788–11789. [Google Scholar]
- Moss, M.; Frey, P.A. The role of S-adenosylmethionine in the lysine 2,3-aminomutase reaction. J. Biol. Chem 1987, 262, 14859–14862. [Google Scholar]
- Baraniak, J.; Moss, M.L.; Frey, P.A. Lysine 2,3-aminomutase. Support for a mechanism of hydrogen transfer involving S-adenosylmethionine. J. Biol. Chem 1989, 264, 1357–1360. [Google Scholar]
- Pieck, J.; Hennecke, U.; Pierik, A.; Friedel, M.; Carell, T. Characterization of a new thermophilic spore photoproduct lyase from geobacillus stearothermophilus (SplG) with defined lesion containing DNA substrates. J. Biol. Chem 2006, 281, 36317–36326. [Google Scholar]
- Benjdia, A.; Heil, K.; Barends, T.R.M.; Carell, T.; Schlichting, I. Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme. Nucleic Acids Res 2012, 40, 9308–9318. [Google Scholar]
- Kneuttinger, A.C.; Heil, K.; Kashiwazaki, G.; Carell, T. The radical SAM enzyme spore photoproduct lyase employs a tyrosyl radical for DNA repair. Chem. Commun 2013, 49, 722–724. [Google Scholar]
- Chandor-Proust, A.; Berteau, O.; Douki, T.; Gasparutto, D.; Ollagnier-De-Choudens, S.; Fontecave, M.; Atta, M. DNA repair and free radicals, new insights into the mechanism of spore photoproduct lyase revealed by single amino Acid substitution. J. Biol. Chem 2008, 283, 36361–36368. [Google Scholar]
- Yang, L.; Lin, G.; Liu, D.; Dria, K.J.; Telser, J.; Li, L. Probing the Reaction Mechanism of Spore Photoproduct Lyase (SPL) via Diastereoselectively Labeled Dinucleotide SP TpT Substrates. J. Am. Chem. Soc 2011, 133, 10434–10447. [Google Scholar]
- Yang, L.; Lin, G.; Nelson, R.S.; Jian, Y.; Telser, J.; Li, L. Mechanistic Studies of the Spore Photoproduct Lyase via a Single Cysteine Mutation. Biochemistry 2012, 51, 7173–7188. [Google Scholar]
- Yang, L.; Nelson, R.S.; Benjdia, A.; Lin, G.; Telser, J.; Stoll, S.; Schlichting, I.; Li, L. A Radical Transfer Pathway in Spore Photoproduct Lyase. Biochemistry 2013, 52, 3041–3050. [Google Scholar]
- Chandra, T.; Silver, S.C.; Zilinskas, E.; Shepard, E.M.; Broderick, W.E.; Broderick, J.B. Spore photoproduct lyase catalyzes specific repair of the 5R but not the 5S spore photoproduct. J. Am. Chem. Soc 2009, 131, 2420–2421. [Google Scholar]
- Silver, S.; Chandra, T.; Zilinskas, E.; Ghose, S.; Broderick, W.; Broderick, J. Complete stereospecific repair of a synthetic dinucleotide spore photoproduct by spore photoproduct lyase. J. Biol. Inorg. Chem 2010, 15, 943–955. [Google Scholar]
- Kim, S.J.; Lester, C.; Begley, T.P. Synthesis of the dinucleotide spore photoproduct. J. Org. Chem 1995, 60, 6256–6257. [Google Scholar]
- Friedel, M.; Berteau, O.; Pieck, J.; Atta, M.; Ollagnier-De-Choudens, S.; Fontecave, M.; Carell, T. The spore photoproduct lyase repairs the 5S- and not the 5R-configured spore photoproduct DNA lesion. Chem. Commun. 2006, 445–447. [Google Scholar]
- Friedel, M.; Pieck, J.; Klages, J.; Dauth, C.; Kessler, H.; Carell, T. Synthesis and stereochemical assignment of DNA spore photoproduct analogues. Chem. Eur. J 2006, 12, 6081–6094. [Google Scholar]
- Mantel, C.; Chandor, A.; Gasparutto, D.; Douki, T.; Atta, M.; Fontecave, M.; Bayle, P.A.; Mouesca, J.M.; Bardet, M. Combined NMR and DFT studies for the absolute configuration elucidation of the spore photoproduct, a UV-induced DNA lesion. J. Am. Chem. Soc 2008, 130, 16978–16984. [Google Scholar]
- Lin, G.; Chen, C.-H.; Pink, M.; Pu, J.; Li, L. Chemical synthesis, crystal structure and enzymatic evaluation of a dinucleotide spore photoproduct analogue containing a formacetal linker. Chem. Eur. J 2011, 17, 9658–9668. [Google Scholar]
- Heil, K.; Kneuttinger, A.C.; Schneider, S.; Lischke, U.; Carell, T. Crystal structures and repair studies reveal the identity and the base-pairing properties of the UV-induced spore photoproduct DNA lesion. Chem. Eur. J 2011, 17, 9651–9657. [Google Scholar]
- Guo, J.; Luo, Y.; Himo, F. DNA repair by spore photoproduct lyase: A density functional theory study. J. Phys. Chem. B 2003, 107, 11188–11192. [Google Scholar]
- Himo, F. C-C bond formation and cleavage in radical enzymes, a theoretical perspective. Biochim. Biophys. Acta 2005, 1707, 24–33. [Google Scholar]
- Fajardo-Cavazos, P.; Rebeil, R.; Nicholson, W. Essential cysteine residues in Bacillus subtilis spore photoproduct lyase identified by alanine scanning mutagenesis. Curr. Microbiol 2005, 51, 331–335. [Google Scholar]
- Lin, G.; Li, L. Oxidation and reduction of 5-(2′-Deoxyuridinyl)methyl radical. Angew. Chem. Int. Ed 2013, 52, 5594–5598. [Google Scholar]
- Mccarty, R.M.; Krebs, C.; Bandarian, V. Spectroscopic, steady-state kinetic, and mechanistic characterization of the radical SAM enzyme QueE, which catalyzes a complex cyclization reaction in the biosynthesis of 7-Deazapurines. Biochemistry 2013, 52, 188–198. [Google Scholar]
- Cotruvo, J.A.; Stich, T.A.; Britt, R.D.; Stubbe, J. Mechanism of assembly of the dimanganese-tyrosyl radical cofactor of class Ib ribonucleotide reductase: Enzymatic generation of superoxide is required for tyrosine oxidation via a Mn(III)Mn(IV) intermediate. J. Am. Chem. Soc 2013, 135, 4027–4039. [Google Scholar]
- Torrents, E.; Sahlin, M.; Biglino, D.; Gräslund, A.; Sjöberg, B.-M. Efficient growth inhibition of Bacillus anthracis by knocking out the ribonucleotide reductase tyrosyl radical. Proc. Natl. Acad. Sci. USA 2005, 102, 17946–17951. [Google Scholar]
- Cotruvo, J.A.; Stubbe, J. Escherichia coli Class Ib ribonucleotide reductase contains a Dimanganese(III)-Tyrosyl radical cofactor in Vivo. Biochemistry 2011, 50, 1672–1681. [Google Scholar]
- Bollinger, J.M., Jr; Edmondson, D.E.; Huynh, B.H.; Filley, J.; Norton, J.R.; Stubbe, J. Mechanism of assembly of the tyrosyl radical-dinuclear iron cluster cofactor of ribonucleotide reductase. Science 1991, 253, 292–298. [Google Scholar]
- Jian, Y.; Li, L. Chemical syntheses of oligodeoxyribonucleotides containing spore photoproduct. J. Org. Chem 2013, 78, 3021–3029. [Google Scholar]

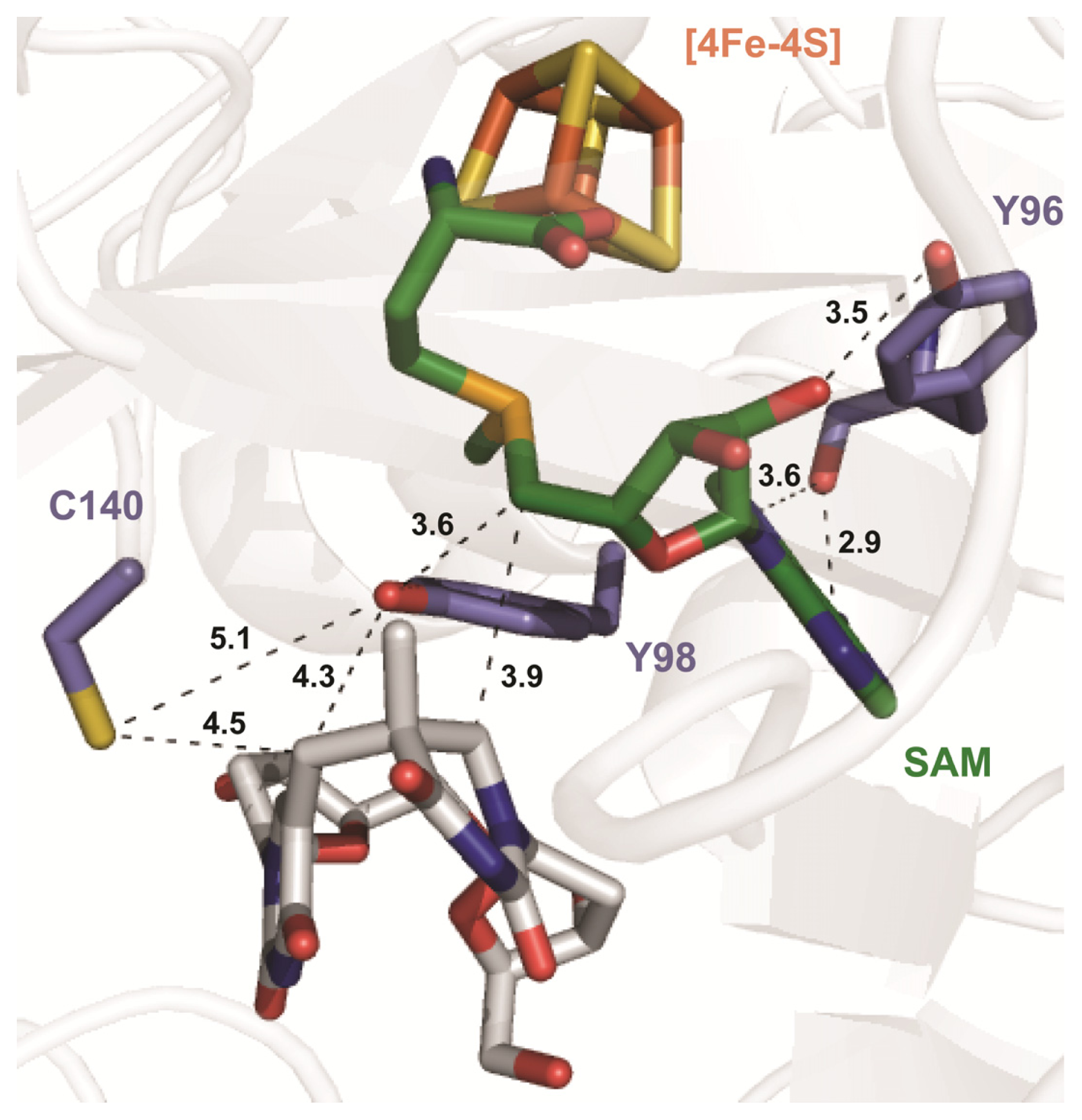
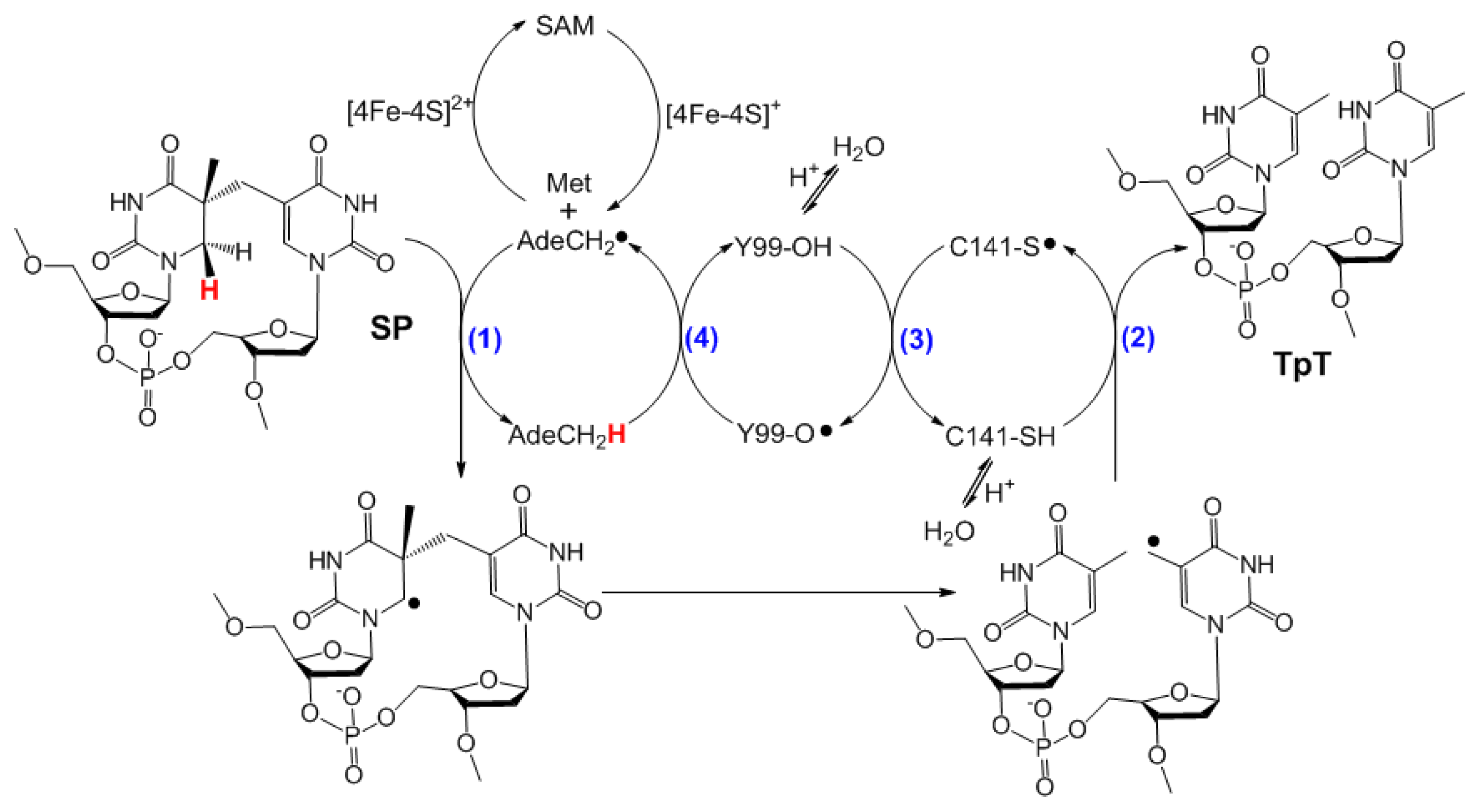
| SPL enzyme | v (min−1) | Apparent KIE | Competitive KIE | SP repaired/5′-dA |
|---|---|---|---|---|
| W.T. | 0.41 ± 0.03 | 2.8 ± 0.3 | 3.4 ± 0.3 | 1.5 ± 0.2 |
| C141A(Bs) | 0.14 ± 0.02 | 1.7 ± 0.2 | 3.0 ± 0.3 | 1.08 ± 0.1 |
| Y97,99A(Bs) | N.A. | N.A. | N.A. | N.A. |
| Y97F(Bs) | 0.12 ± 0.01 | 16 ± 1.5 | 11.5 ± 1.5 | 1.6 ± 0.2 |
| Y99F(Bs) | 0.06 ± 0.005 | 10.5 ± 1 | 9 ± 1 | 1.0 ± 0.1 |
| Y97,99F(Bs) | <0.004 | N.A. | N.A. | 0.92 ± 0.1 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yang, L.; Li, L. The Enzyme-Mediated Direct Reversal of a Dithymine Photoproduct in Germinating Endospores. Int. J. Mol. Sci. 2013, 14, 13137-13153. https://doi.org/10.3390/ijms140713137
Yang L, Li L. The Enzyme-Mediated Direct Reversal of a Dithymine Photoproduct in Germinating Endospores. International Journal of Molecular Sciences. 2013; 14(7):13137-13153. https://doi.org/10.3390/ijms140713137
Chicago/Turabian StyleYang, Linlin, and Lei Li. 2013. "The Enzyme-Mediated Direct Reversal of a Dithymine Photoproduct in Germinating Endospores" International Journal of Molecular Sciences 14, no. 7: 13137-13153. https://doi.org/10.3390/ijms140713137




