1. Introduction
The heart is the first organ to form and function during vertebrate embryonic development. Congenital heart disease is an important component of pediatric cardiovascular disease present at birth, and constitutes a major percentage of clinically significant birth defects, with an estimated prevalence of 4–50 per 1000 live births [
1,
2]. Congenital heart defects occur in almost 1% of live births, reflecting the complex cellular processes underlying heart development [
3]. Cardiac development requires a precise and extremely complex series of molecular mechanisms to orchestrate the proper expression of cardiac transcription factors, and alterations in their expression can result in heart defects [
4,
5]. Classical forward genetic human pedigree studies have thus far provided limited insight into the genetic causes of congenital heart defects.
Previously, McCann
et al. found that fatty acid-binding protein 3 (FABP3; also referred to as heart-type fatty acid-binding protein) could be used as a novel plasma biomarker in the early diagnosis of acute ischemic chest pain [
6]. Earlier, our group also established that FABP3 is upregulated in patients with ventricular-septal defects in comparison to normal controls, by using subtractive hybridization [
7]. We have also reported that down-regulation of FABP3 expression promotes cell apoptosis and causes mitochondrial dysfunction in P19 cells [
8]. Interestingly, FABP3 has been shown to be upregulated during terminal differentiation of mouse cardiomyocytes [
9]. Taken together these studies suggest that FABP3 may play an important role in cardiac development; however, definitive data is lacking.
One factor that is known to be important in cardiac development is retinoic acid (RA). For example, the size of the myocardial progenitor pool is restricted by RA signaling [
10], and evidence from several studies has suggested that excess or reduced levels of RA can result in cardiac defects [
10,
11]. It is known that altered RA levels by regulated activation of the Raldh2 gene in avian embryos supports a role for RA signaling in development of the venous pole of the heart [
12].
In recent years, the zebrafish has become one of the best vertebrate models for studying developmental biology. Cardiovascular development in the zebrafish is well defined and has been shown to be morphologically and physiologically similar to that of mammals [
13,
14]. Additionally, early-stage cardiac development of zebrafish is similar to that of the human heart in many respects, such as migration of cardiac precursor cells towards the central line, heart tube formation, early chamber formation and the looping process [
13,
15]. Thus, studies in the zebrafish embryo can facilitate the characterization of cardiac defects and their underlying molecular mechanisms.
As discussed above, FABP3 is a gene of interest in cardiac development; moreover, it has previously been established that FABP3 strong expressed in the central nervous system (CNS), retina and myotomes using situ hybridization, FABP3 mRNA is expressed in the developing zebrafish [
16,
17]. In this study we used a zebrafish model to investigate the role of FABP3 in cardiac development and dissect its underlying molecular mechanisms, particularly with regard to its interaction with the RA signaling pathway. To achieve this goal, we used a morpholino (MO) approach to silence FABP3 expression and then examined the resultant effect of FABP3 knockdown on cardiac development and heart function in zebrafish embryos. We also analyzed the alteration of RA signaling pathway to determine if FABP3 knockdown has an effect on this important pathway in cardiac development.
3. Discussion
In the last decade, zebrafish have been used as a powerful model to study cardiac development [
13]. The mechanisms of zebrafish development are thought to be similar to mammals, thus an improved understanding of zebrafish development can be related to developmental mechanisms in these animals. The zebrafish embryonic attributes augment the feasibility of conducting classical genetic screens that affect cardiac development. Thus, we used zebrafish as a model to investigate the FABP3 in cardiac development. We have previously reported that FABP3 is highly expressed in patients with ventricular-septal defects, when compared with normal controls [
7]. This and other observations led us to hypothesize that FABP3 might play an important role in cardiac development. In the present study, cardiac development was explored in FABP3-MO injected zebrafish embryos and the resultant zebrafish heart defects were further studied at the molecular level.
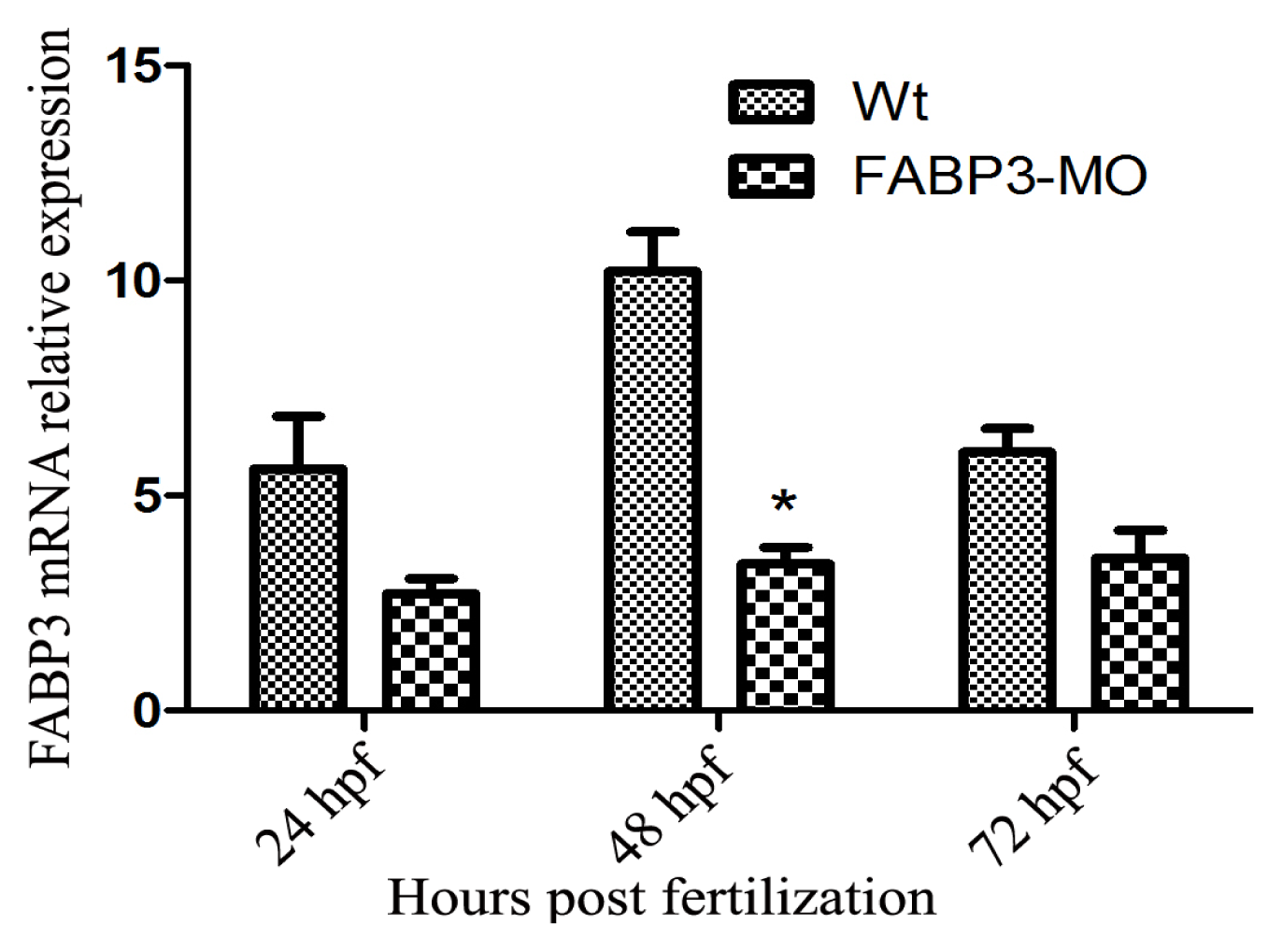
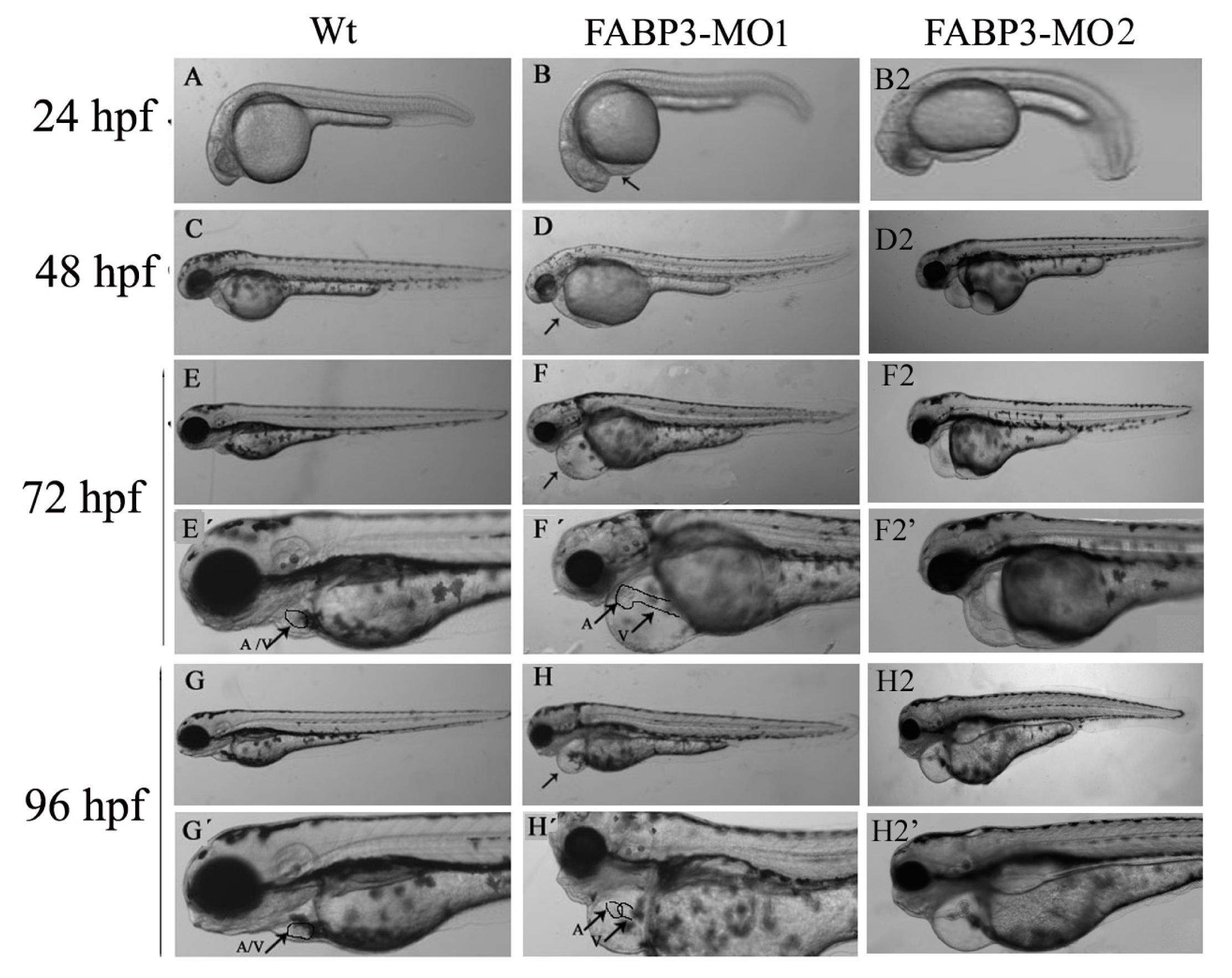
After establishing that our FABP3-MO led to knockdown of FABP3 in zebrafish embryos, we performed a systematic investigation of zebrafish heart development. We found that injection of FABP3-MO substantially increased the rate of embryonic lethality in a dose-dependent manner. Based on these preliminary studies, a dose of 5.0 ng of FABP3-MO was chosen to study the effect of FABP3 knockdown in heart development. To minimize off-target effects and strengthen the impact of the observed heart phenotype, we designed the splice-Morpholino of FABP3 (FABP3-MO2) and observed the heart phenotype. At a dose of 5.0 ng MO, we found embryonic or larvae malformations, developmental delay, smaller heads, abnormal eye development, pericardial edema and a linear heart tube phenotype in FABP3-MO injected zebrafish. We interpreted these results to indicate that FABP3 can influence the development of zebrafish, especially with regard to the heart.
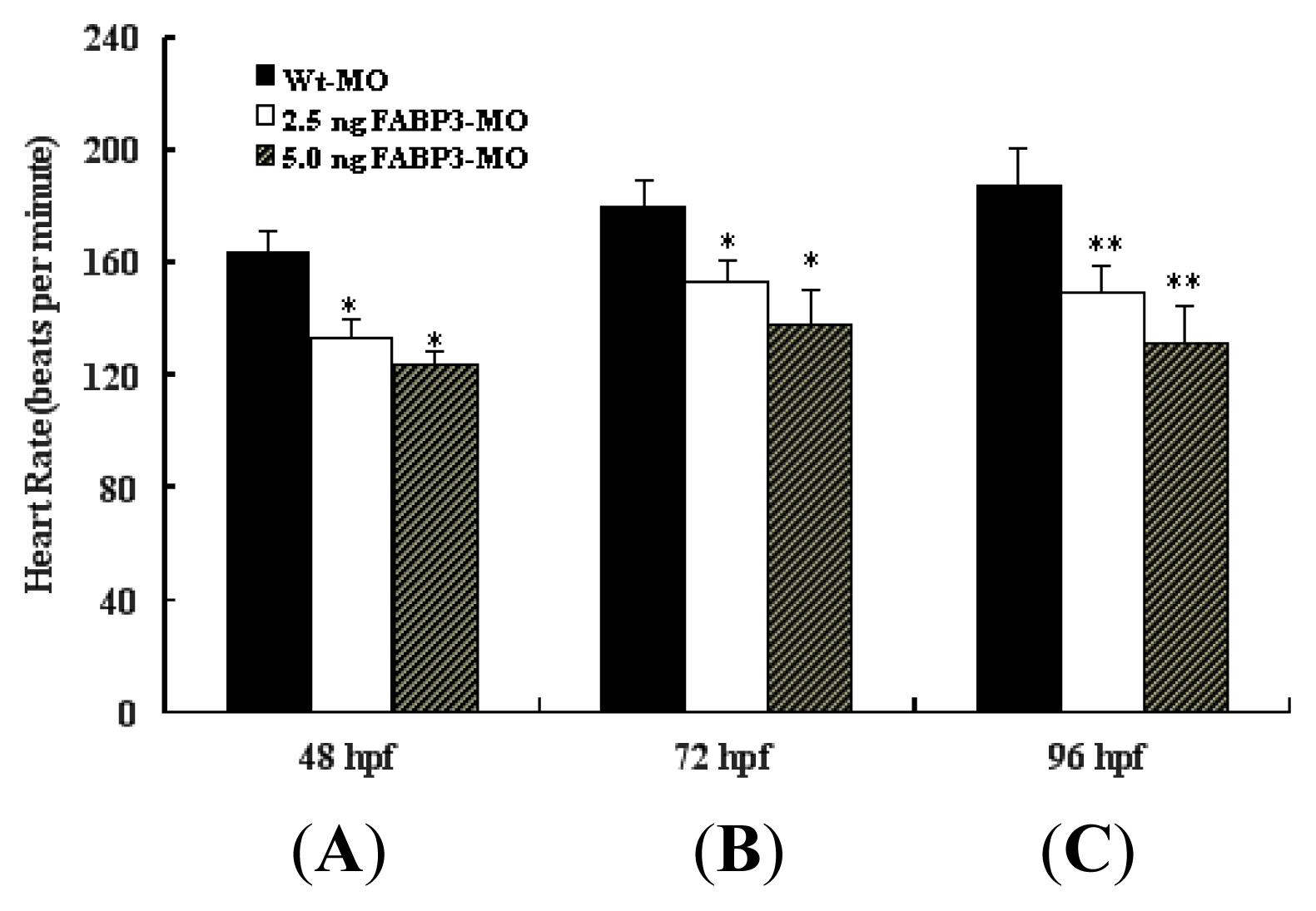
It is known that when the bilateral heart fields fuse at the mid-line, they form a cardiac disc structure with the endocardial cells within a hole at the center, atrial myocytes at the periphery and ventricular myocytes around the circumference of the disc [
15]. After heart tube fusion, the heart begins to slowly beat at about 25 times/min at 22 hpf, then the heartbeat gradually increases to about 90 times/min. Interestingly, in addition to the structural defects described above, decreased heart rate was also detected in FABP3-MO injected embryos; however, the trajectory of heart rate was increased in FABP3-MO injected embryos from 48 to 96 hpf (
Figure 3). This data confirmed that FABP3-MO injection impaired heart development, which was reflected in decreased heart function.
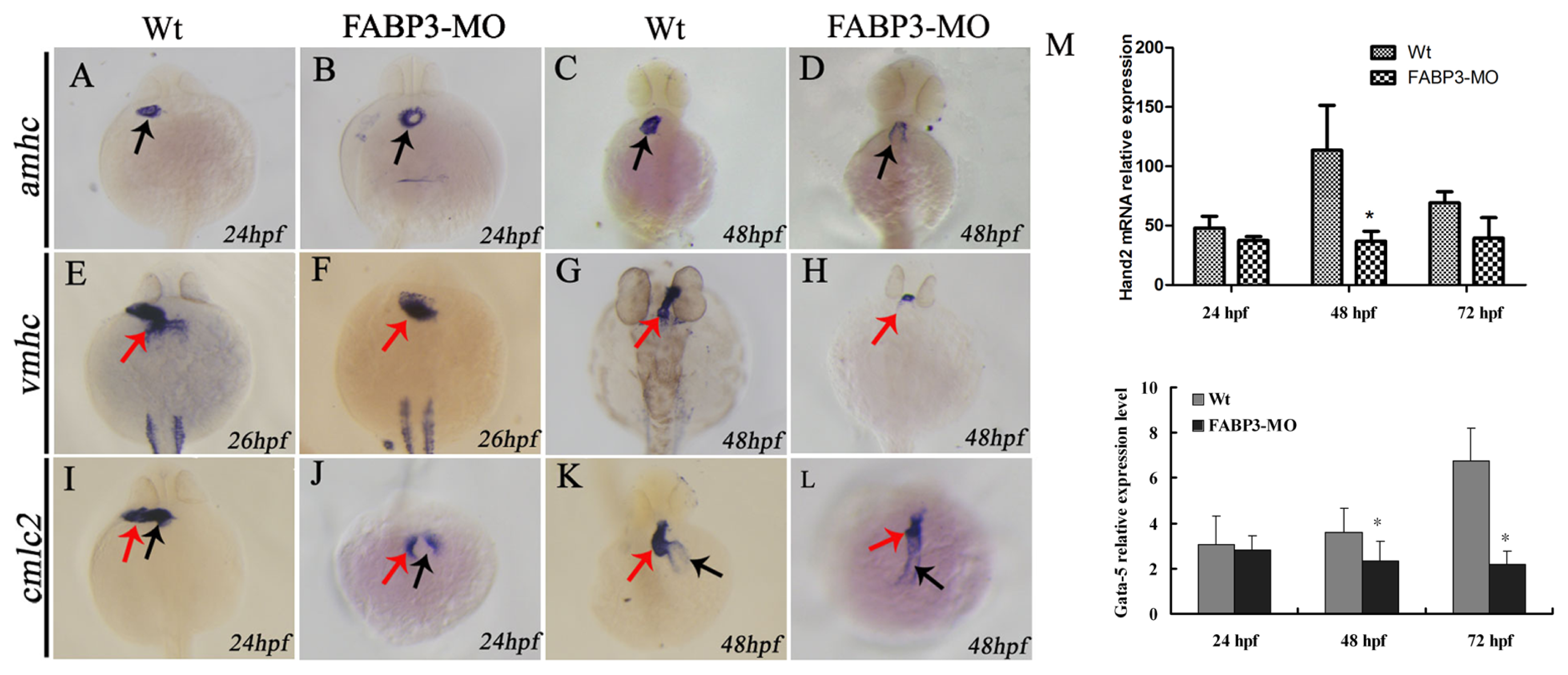
The myocardial cell population of the developing heart is polarized in a medial to lateral direction, with the medial cells expressing vmhc and the lateral cells expressing amhc. Expression of amhc is known to be initiated slightly later compared than the onset of expression of vmhc [
22], which suggested that the differentiation of these two myocardial cell types is initiated at different time points. Expression of cmlc2 is initiated in a few cells, with the number of cells expressing cmlc2 increasing over time [
20]. This strict organization and separation of ventricular and atrial myocytes is maintained during the later stages of cardiac development, although it is currently unclear by what mechanism these two cell populations are kept separated. Given the importance of these factors in heart development, we analyzed their expression level in FABP3-MO injected embryonic zebrafish, with our results showing that amhc and vmhc expression was downregulated. Furthermore, our cmlc2 expression data indicated that heart looping was incomplete in FABP3-MO injected zebrafish, which was consistent with our morphological examination of the developing heart. Taken together, our data strongly indicated that FABP3-MO injection impaired cardiac development in zebrafish embryos, and that these hearts underwent abnormal heart looping.
It is known that Hand2 and Gata5 are required for cardiogenic differentiation, because hands off/hand2 mutant embryos have dramatically reduced myocardial tissue [
23]. In addition, Gata5 interact to induce Hand2 expression. It has also been established that Hand2 also has a non-cell-autonomous role during fusion of the bilateral cardiac fields by repressing fibronectin production [
24]. Our results showed that Hand2 and Gata5 are downregulated in zebrafish injected with FABP3-MO at 48 and 72 hpf, and this time point was also the FABP3 expression level decreased when zebrafish injected with FABP3-MO, indicating that FABP3 may be an upstream regulator of Gata5 and Hand2, and that the heart defects observed in FABP3-MO injected embryos may have their origin in dysregulated Gata5 and Hand2 expression. However, the time course of FABP3 affected betweenGata5 and Hand2 expression was still unknown, which needs further study.
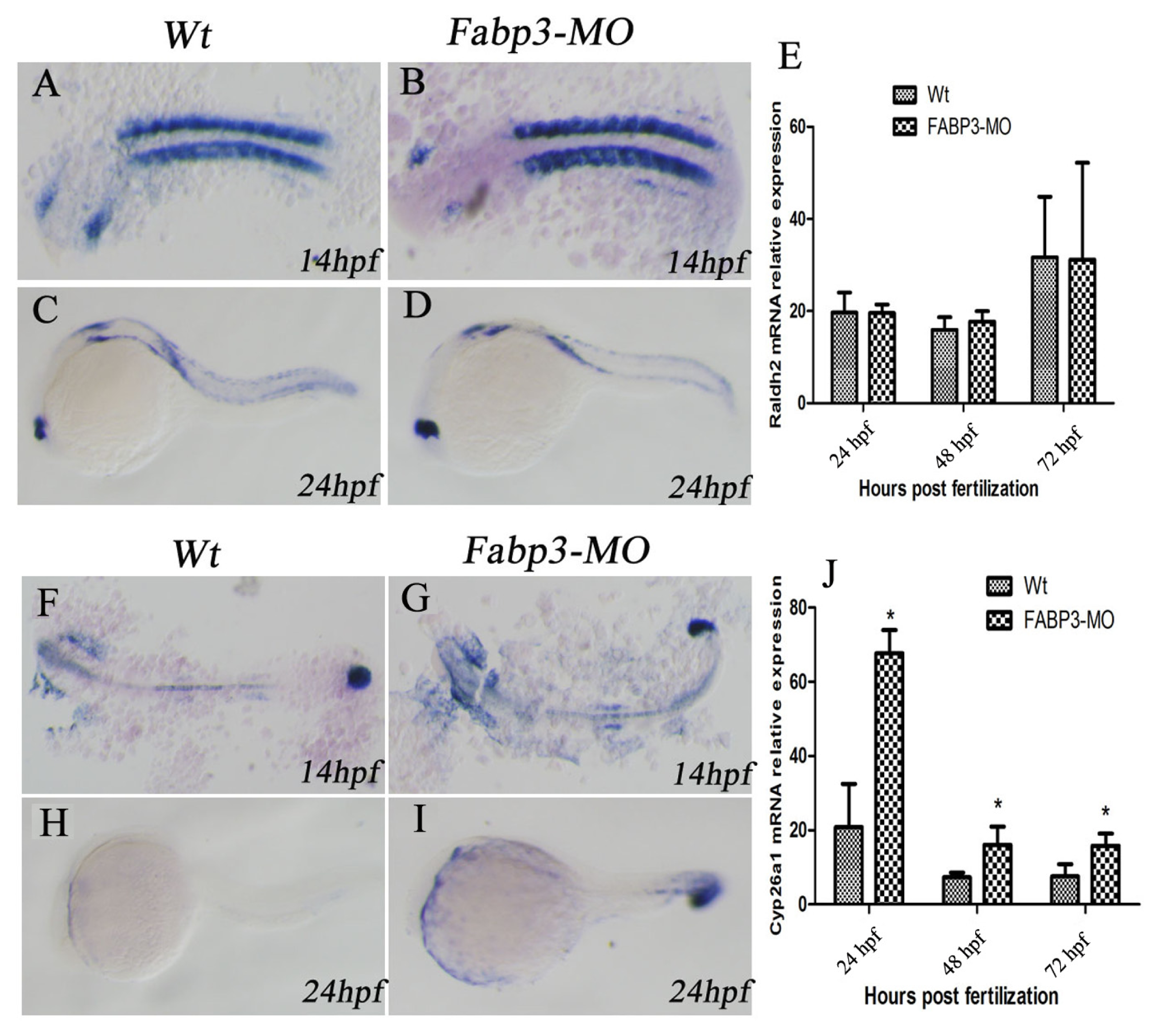
In addition to the important heart genes, we were also interested in examining other signaling pathways that are known to be important in heart development. In this regard, it is known that multiple signaling pathways converge to regulate heart development, including RA, Wnt, BMP, and Hh signaling [
25]. In this study, we focused on the RA signaling pathway in FABP3-MO injected zebrafish. Our results indicated that the RA-metabolizing enzyme, Cyp26a1 was upregulated in FABP3-MO injected embryos, though there was no significant effect on the expression of Raldh2, which synthesizes RA. It is known that RA can regulate the size of the cardiac field in zebrafish [
10,
12], and that excess or reduced RA levels can result in cardiac defects. For example, altered RA levels in avian embryos supports a role for RA signaling in development of the venous pole of the heart [
12]. It has also recently been suggested that RA signaling may participate in defining the posterior limit of the second heart field formation, heart tube fails to extend in Raldh2 mutant embryos [
11,
26]. Interestingly, knockout of Raldh2 in mouse embryos reveals that a number of second heart field formation genes are abnormally expanded in a posterior direction, including Isl1, Tbx1, Fgf10, and Fgf8 [
11,
26]. Besides, retinoic acid signaling is in turn controlled by signals from the second heart field: Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function [
27] Our results indicated that FABP3-MO influences the RA signaling pathway through altered Cyp26a1 expression, which could lead to cardiac developmental abnormalities by dysregulating RA levels in the developing heart.
4. Experimental Section
4.1. Embryo Maintenance and Transplantation
Wild type zebrafish stocks were obtained from the Model Animal Research Center of Nanjing University. Embryos were obtained from natural spawning of wild type adults, and were grown at 28.5 °C in embryo medium as previously described [
28]. Morphological features were used to determine the embryonic developmental stage, as described by Kimmel
et al. [
29]. Embryos older than 24 h post-fertilization (hpf) were incubated in 0.003% phenylthiourea to inhibit pigment formation.
4.2. Morpholinos and Microinjection
MOs (morpholinos) were purchased from Gene Tools (Philomath, OR, USA). The FABP3 MO was designed against the ATG start site of FABP3 to block its translation (FABP3-MO1; 5′-ACG TGC CGA TAA AAG CGT CTG CCA T-3′), and we also designed the oligo to target the exon 2/intron 2 splice boundary by Gene Tools (FABP3-MO2; 5′-AGC CAA CAC CTG ATA AAG CAA ACA T-3′). A standard control MO (Wt-MO; 5′-CCT CTT ACC TCA GTT ACA ATT TAT A-3′) served as negative control. MO oligomers were dissolved in 0.3 × Danieaus’s solution (58 mmol/L NaCl, 0.7 mmol/L KCl, 0.4 mmol/L MgSO
4, 0.6 mmol/L Ca(NO
3)
2, 5.0 mmol/L
N-[2-Hydroxyethyl] piperazine-
N′-[2-ethanesulfonic acid] (HEPES), pH7.6) at 2 mmol/L [
20], and diluted to their final concentration in 0.2 mol/L KCl and 0.5% (
w/
v) phenol red (Sigma, St. Louis, MO, USA). Injection of 1.0, 2.5, 5.0, 7.5 and 10.0 ng of MO into single to four-cell stage zebrafish embryos using back-filled fine borosilicate glass capillary needles was performed as previously described [
18]. Following MO injection, embryos were incubated at 28.5 °C for up to 3 days post-fertilization.
4.3. Determining the Rate of Lethality and Observation of Zebrafish Heart Development
After microinjection of MOs into zebrafish embryos, we counted the rates of lethality and identified heart-specific phenotypes and malformations using an Olympus SZ61 dissecting microscope (Olympus, Tokyo, Japan) at the following time points: 24, 48, 72 and 96 hpf. Embryos were photographed with a DP70 digital camera (Olympus), and images were processed using Adobe Photoshop software (Adobe Systems, San Jose, CA, USA).
4.4. Heart Rate Measurement
Embryos were anesthetized in ethyl aminoboenzoate (Tricaine, Sigma, St. Louis, MO, USA). A Tricaine solution stock was made as follows: 400 mg of Tricaine powder, 97.9 mL ddH2O and 2.1 mL 1 mol/L Tris (pH 9). The solution was adjusted to pH 7 and stored in the freezer until required for further use. To anesthetize fish, 4.2 mL of Tricaine solution stock was diluted in a clean 100 mL water tank. Anesthetized embryos were then transferred to a recording chamber perfused with modified Tyrode’s solution (136 mmol/L NaCl, 5.4 mmol/L KCl, 0.3 mmol/L NaH2PO4, 1.8 mmol/L CaCl2, 1 mmol/L MgCl2, 10 mmol/L HEPES, 5 mmol/L glucose, pH 7.3) at 48 hpf. The heart rate was calculated by counting the number of sequential contractions, beginning and ending at the end diastole. This was conducted under a dissecting microscope in 30 s intervals.
4.5. Preparation of RNA Probes
RNA Probes used for in situ hybridization included: atrial myosin heavy chain (amhc), ventricula rmyosin heavy chain (vmhc), cardiacmyosin light chain-2 (cmlc2), retinaldehyde dehydrogenase gene (Raldh2) and Cytochrome P450 26A1 (Cyp26a1). Probes were amplified from cDNA generated from embryos at 48 hpf and subcloned into pBluescript-KS vector (Stratagene, La Jolla, CA, USA). These plasmids were then linearized and transcribed as follows: amhc, SalI (Takara, Osaka, Japan)/SP6; vmhc and cmlc2, SalI/SP7; Raldh2 and Cyp26a1, NcoI (Takara, Osaka, Japan))/SP6. Each RNA probe was transcribed using a MAXIscript®In Vitro Transcription Kit (Invitrogen, Carlsbad, CA, USA). Digoxigenin-labeled single strand RNA probes were prepared using a digoxigenin RNA labeling kit (Roche Diagnostics, Basle, Switzerland) according to the manufacturer’s instructions, and stored at −40 °C for future use. For each gene, a sense (forward) probe was used as control, but was found to yield no specific signal.
4.6. Whole-Mount in Situ Hybridization
RNA in situ hybridization was performed as previously described [
30] using riboprobes specific for vmhc, amhc, cmlc2, Raldh2 and Cyp26a1 [
31]. To perform whole-mount in situ hybridization, we fixed the embryos overnight with PBS-based 4% paraformaldehyde at 4 °C. The following day, embryos were dechorionated manually and then transferred to 1.5 mL Eppendorf tubes. The embryos were dehydrated in a graded series of 50% methanol/PBS, 75% methanol/PBS and 100% methanol for 5 min, respectively. The 100% methanol was changed twice at 5 min intervals, and then the embryos were stored at −20 °C for at least 3 h until required for further processing. All further steps were performed at room temperature unless stated otherwise. To continue the experiment, the embryos were rehydrated by incubating for 5 min each in 50% methanol in PBST (PBS/0.1% Tween-20), 25% methanol in PBST, and twice in PBST. Embryos were then incubated at 65 °C for 5 min in tubes containing Hyb-solution (50% formamide, 5× SSC, 0.1% Tween-20). The Hyb-solution was replaced by Hyb+ (Hyb− with 5 mg/mL yeast RNA [Sigma, St. Louis, MO, USA]; and 50 mg/mL heparin [Sigma]) and the embryos were prehybridized in Hyb+ at 65 °C for 4 h up to 24 h. As much of the Hyb+ solution as possible was removed without allowing the embryos to come in contact with air, before 100 μL of fresh Hyb+ containing 50 to 100 ng of digoxigenin-labeled RNA probe (heated to 65 °C for 10 min then on ice 2 min prior to addition) was added to the tubes. After an overnight incubation at 65 °C the probe was removed and the embryos were washed twice for 30 min in 50% formamide/2 × SSC at 65 °C, followed by washing with 2× SSC (15 min at 65 °C) and two washes with 0.2× SSCT (30 min at 65 °C). Then embryos were blocked for 1 h in blocking buffer (2% blocking reagent [Roche, Indianapolis, IN, USA], 10% sheep serum and 70% Maleic acid (MAB) [Sigma, St. Louis, MO, USA]). Alkaline-phosphatase conjugated anti-digoxigenin fab-fragments (1:5000, Roche) were added in fresh blocking buffer. After overnight incubation, embryos were washed 8 times with MABT (MAB/0.1% Tween-20) (45 min each). Then, the overnight MABT washing solution was changed with equilibration buffer (100 mM Tris pH 9.5, 50 mM MgCl
2, 100 mM NaCl, 1 mM levamisol [Sigma], and 0.1% Tween-20), and incubated with the buffer for 10 min. Detection was performed with new equilibration buffer containing the substrate NBT/BCIP solution. The reaction was monitored under a dissecting microscope and stopped when the hybridization signal was satisfactory. Stained specimens were washed in PBST for 5 min, dehydrated in methanol twice for 10 min each and mounted in 2:1 benzyl benzoate: benzyl alcohol (Sigma). Photomicrographs were taken using an Olympus DP71 digital camera (Olympus), and digital images were further processed for brightness and contrast with Adobe Photoshop software (version 7.0; Adobe Systems: San Jose, CA, USA).
4.7. Real-Time Polymerase Chain Reaction (PCR) for the Assessment of Cardiac-Specific Gene Expression
Total RNA was extracted from zebrafish embryos using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and the extracted RNA was quantified using a NanoDrop 2.0 spectrophotometer (Thermo Scientific; Waltham, MA, USA). Complimentary DNA was synthesized from 2 μg of total RNA by using an AMV Reverse Transcriptase Kit (Promega A3500; Promega, Madison, WI, USA) according to the manufacturer’s instructions. Real-time qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) on an Applied Biosystems 7500 Sequence Detection System (ABI 7500 SDS). Real-time PCR reaction conditions were 95 °C for 10 min followed by 40 cycles of 94 °C for 20 s, 60 °C for 20 s, and 72 °C for 30 s with a final extension of 7 min at 72 °C. FABP3, helix-loop-helix transcription factor (Hand2), Gata5, Raldh2 and Cyp26a1 expression was normalized against β-actin using the comparative CT method to determine the relative changes in mRNA expression in the target sample. The sequences of the primers used for each gene are shown in the
Table 2.
4.8. Statistical Analysis
All values for statistical significance represent the mean ± standard deviation (SD). Statistical analyses were performed using the chi-squared test for categorical data (SPSS version 16.0; IBM Corporation: Armonk, NY, USA); dose-response relationships were analyzed by the Mantel–Haenszel test. Differences were considered significant for p < 0.05. Means and standard deviations were determined for at least three independent experimental replicates.