Intercellular Communication by Exosome-Derived microRNAs in Cancer
Abstract
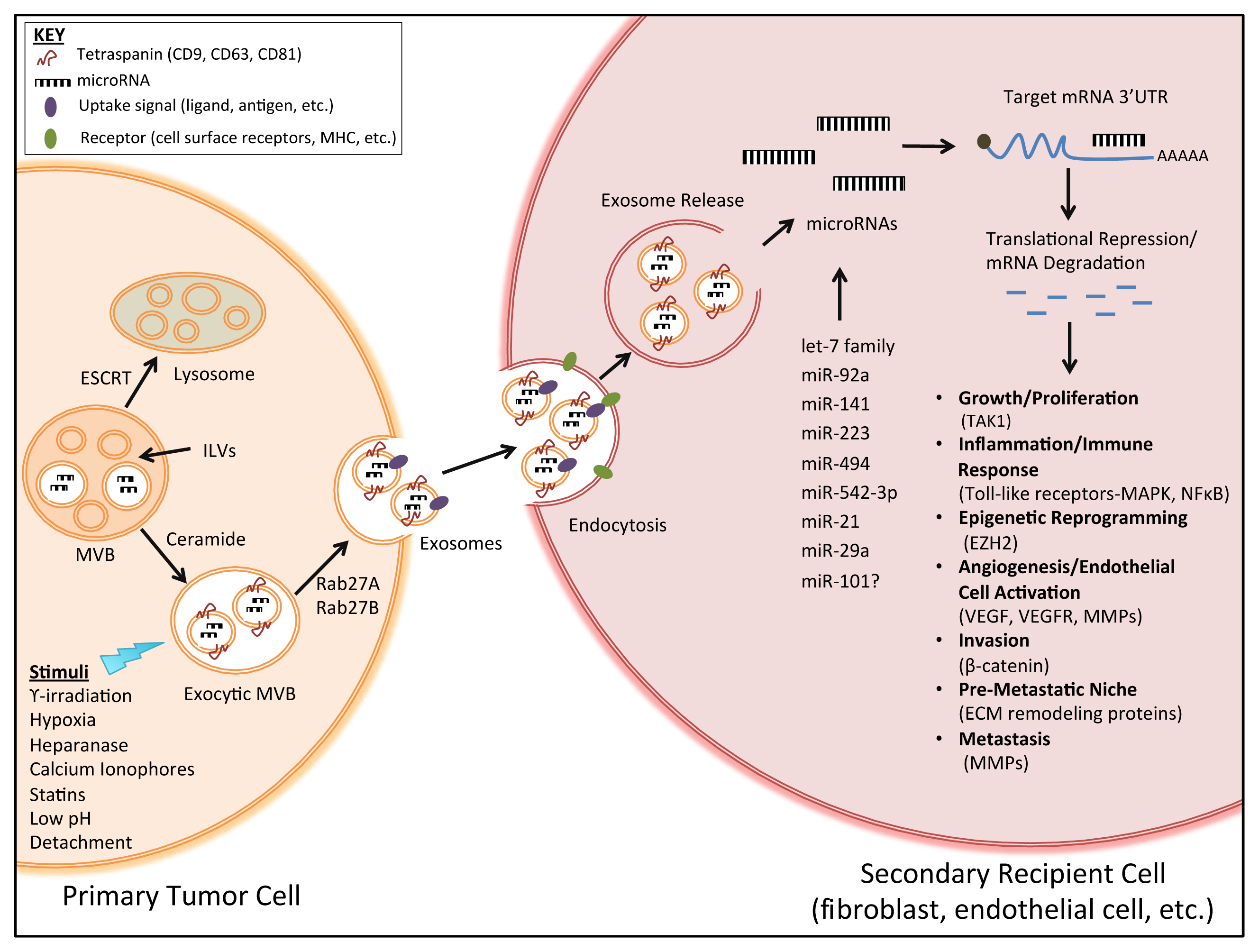
:1. Introduction
1.1. Historical Perspective
1.2. Exosome Biogenesis and Secretion
1.3. Exosome Components
1.4. Exosome Isolation and Examination
2. Exosomes and Intercellular Communication in Tumor Progression
2.1. Exosome Mediation of Intercellular Communication
2.2. Exosome Modulation of Extracellular Matrix, Stromal Cells and Invasion
2.3. Exosome Stimulation of Tumor Angiogenesis and Metastatic Niche Formation
3. Exosome-Derived microRNAs in Tumor Progression
3.1. Exosomes as Transporters of microRNAs
3.2. Secretion and Uptake of Exosome-Derived microRNAs
3.3. microRNA Profiling of Tumor-Derived Exosomes in Clinical Samples
3.4. Pro-Tumorigenic Effects of Exosome-Derived microRNAs in Vitro
4. Exosomes and Breast Cancer
4.1. Exosomes in Normal Mammary Epithelium
4.2. Exosomes in Breast Cancer Development and Progression
4.3. Potential for Diagnostic, Prognostic and Therapeutic Interference
5. Future Directions
Acknowledgments
Conflict of Interest
References
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol 2002, 2, 569–579. [Google Scholar]
- Wickman, G.; Julian, L.; Olson, M.F. How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ 2012, 19, 735–742. [Google Scholar]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol 1985, 101, 942–948. [Google Scholar]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol 1983, 97, 329–339. [Google Scholar]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol 2007, 179, 1969–1978. [Google Scholar]
- Masyuk, A.I.; Masyuk, T.V.; Larusso, N.F. Exosomes in the pathogenesis, diagnostics and therapeutics of liver diseases. J. Hepatol. 2013. [Google Scholar] [CrossRef]
- Vella, L.J.; Sharples, R.A.; Nisbet, R.M.; Cappai, R.; Hill, A.F. The role of exosomes in the processing of proteins associated with neurodegenerative diseases. Eur. Biophys. J 2008, 37, 323–332. [Google Scholar]
- Keller, S.; Rupp, C.; Stoeck, A.; Runz, S.; Fogel, M.; Lugert, S.; Hager, H.D.; Abdel-Bakky, M.S.; Gutwein, P.; Altevogt, P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int 2007, 72, 1095–1102. [Google Scholar]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol 2008, 110, 13–21. [Google Scholar]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 2012, 7, e30679. [Google Scholar]
- Saman, S.; Kim, W.; Raya, M.; Visnick, Y.; Miro, S.; Saman, S.; Jackson, B.; McKee, A.C.; Alvarez, V.E.; Lee, N.C.; et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J. Biol. Chem 2012, 287, 3842–3849. [Google Scholar]
- Qiu, S.; Duan, X.; Geng, X.; Xie, J.; Gao, H. Antigen-specific activities of CD8+ T cells in the nasal mucosa of patients with nasal allergy. Asian Pac. J. Allergy Immunol. Launched Allergy Immunol. Soc. Thail 2012, 30, 107–113. [Google Scholar]
- Record, M. Exosomal Lipids in Cell–Cell Communication. In Emerging Concepts of Tumor Exosome–Mediated Cell-Cell Communication; Zhang, H.-G., Ed.; Springer: New York, NY, USA, 2013; pp. 47–68. [Google Scholar]
- Bobrie, A.; Colombo, M.; Raposo, G.; Thery, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar]
- Hanson, P.I.; Cashikar, A. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol 2012, 28, 337–362. [Google Scholar]
- Schmidt, O.; Teis, D. The ESCRT machinery. Curr. Biol 2012, 22, R116–R120. [Google Scholar]
- Raymond, C.K.; Howald-Stevenson, I.; Vater, C.A.; Stevens, T.H. Morphological classification of the yeast vacuolar protein sorting mutants: Evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 1992, 3, 1389–1402. [Google Scholar]
- Raiborg, C.; Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009, 458, 445–452. [Google Scholar]
- Katzmann, D.J.; Babst, M.; Emr, S.D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001, 106, 145–155. [Google Scholar]
- Wollert, T.; Wunder, C.; Lippincott-Schwartz, J.; Hurley, J.H. Membrane scission by the ESCRT-III complex. Nature 2009, 458, 172–177. [Google Scholar]
- Van Blitterswijk, W.J.; van der Luit, A.H.; Veldman, R.J.; Verheij, M.; Borst, J. Ceramide: Second messenger or modulator of membrane structure and dynamics? Biochem. J 2003, 369, 199–211. [Google Scholar]
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212. [Google Scholar]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar]
- Van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar]
- Rodriguez-Boulan, E.; Kreitzer, G.; Musch, A. Organization of vesicular trafficking in epithelia. Nat. Rev. Mol. Cell Biol 2005, 6, 233–247. [Google Scholar]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev 2011, 91, 119–149. [Google Scholar]
- Savina, A.; Vidal, M.; Colombo, M.I. The exosome pathway in K562 cells is regulated by Rab11. J. Cell Sci 2002, 115, 2505–2515. [Google Scholar]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol 2010, 12, 19–30. [Google Scholar]
- Yu, X.; Harris, S.L.; Levine, A.J. The regulation of exosome secretion: A novel function of the p53 protein. Cancer Res 2006, 66, 4795–4801. [Google Scholar]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [Green Version]
- Thompson, C.A.; Purushothaman, A.; Ramani, V.C.; Vlodavsky, I.; Sanderson, R.D. Heparanase Regulates Secretion, Composition, and Function of Tumor Cell-derived Exosomes. J. Biol. Chem 2013, 288, 10093–10099. [Google Scholar]
- Koumangoye, R.B.; Sakwe, A.M.; Goodwin, J.S.; Patel, T.; Ochieng, J. Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. PLoS One 2011, 6, e24234. [Google Scholar]
- Llorente, A.; van Deurs, B.; Sandvig, K. Cholesterol regulates prostasome release from secretory lysosomes in PC-3 human prostate cancer cells. Eur. J. Cell Biol 2007, 86, 405–415. [Google Scholar]
- Faure, J.; Lachenal, G.; Court, M.; Hirrlinger, J.; Chatellard-Causse, C.; Blot, B.; Grange, J.; Schoehn, G.; Goldberg, Y.; Boyer, V.; et al. Exosomes are released by cultured cortical neurones. Mol. Cell. Neurosci 2006, 31, 642–648. [Google Scholar]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; de Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem 2009, 284, 34211–34222. [Google Scholar]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun 2011, 2, 282. [Google Scholar]
- Keller, S.; Konig, A.K.; Marme, F.; Runz, S.; Wolterink, S.; Koensgen, D.; Mustea, A.; Sehouli, J.; Altevogt, P. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett 2009, 278, 73–81. [Google Scholar]
- Barres, C.; Blanc, L.; Bette-Bobillo, P.; Andre, S.; Mamoun, R.; Gabius, H.J.; Vidal, M. Galectin-5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood 2010, 115, 696–705. [Google Scholar]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res 2012, 40, D1241–D1244. [Google Scholar]
- Mathivanan, S.; Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9, 4997–5000. [Google Scholar]
- Subra, C.; Grand, D.; Laulagnier, K.; Stella, A.; Lambeau, G.; Paillasse, M.; de Medina, P.; Monsarrat, B.; Perret, B.; Silvente-Poirot, S.; et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res 2010, 51, 2105–2120. [Google Scholar]
- Zakharova, L.; Svetlova, M.; Fomina, A.F. T cell exosomes induce cholesterol accumulation in human monocytes via phosphatidylserine receptor. J. Cell. Physiol 2007, 212, 174–181. [Google Scholar]
- Xiang, X.; Poliakov, A.; Liu, C.; Liu, Y.; Deng, Z.B.; Wang, J.; Cheng, Z.; Shah, S.V.; Wang, G.J.; Zhang, L.; et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int. J. Cancer 2009, 124, 2621–2633. [Google Scholar]
- Cao, W.; Ma, Z.; Rasenick, M.M.; Yeh, S.; Yu, J. N-3 poly-unsaturated fatty acids shift estrogen signaling to inhibit human breast cancer cell growth. PLoS One 2012, 7, e52838. [Google Scholar]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 2007, 9, 654–659. [Google Scholar]
- Hessvik, N.P.; Phuyal, S.; Brech, A.; Sandvig, K.; Llorente, A. Profiling of microRNAs in exosomes released from PC-3 prostate cancer cells. Biochim. Biophys. Acta 2012, 1819, 1154–1163. [Google Scholar]
- Bellingham, S.A.; Coleman, B.M.; Hill, A.F. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res 2012, 40, 10937–10949. [Google Scholar]
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar]
- Pigati, L.; Yaddanapudi, S.C.; Iyengar, R.; Kim, D.J.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One 2010, 5, e13515. [Google Scholar]
- Jaiswal, R.; Luk, F.; Gong, J.; Mathys, J.M.; Grau, G.E.; Bebawy, M. Microparticle conferred microRNA profiles—Implications in the transfer and dominance of cancer traits. Mol. Cancer 2012, 11, 37. [Google Scholar]
- Xiao, D.; Ohlendorf, J.; Chen, Y.; Taylor, D.D.; Rai, S.N.; Waigel, S.; Zacharias, W.; Hao, H.; McMasters, K.M. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PLoS One 2012, 7, e46874. [Google Scholar]
- Simpson, R.J.; Jensen, S.S.; Lim, J.W. Proteomic profiling of exosomes: Current perspectives. Proteomics 2008, 8, 4083–4099. [Google Scholar]
- de Gassart, A.; Geminard, C.; Fevrier, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006. [Google Scholar] [CrossRef]
- Cantin, R.; Diou, J.; Belanger, D.; Tremblay, A.M.; Gilbert, C. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. J. Immunol. Methods 2008, 338, 21–30. [Google Scholar]
- Lamparski, H.G.; Metha-Damani, A.; Yao, J.Y.; Patel, S.; Hsu, D.H.; Ruegg, C.; Le Pecq, J.B. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods 2002, 270, 211–226. [Google Scholar]
- System Biosciences. Available online: http://www.systembio.com/exoquick (on accessed 15 April 2013).
- Taylor, D.D.; Atay, S.; Metzinger, D.S.; Gercel-Taylor, C. Characterization of humoral responses of ovarian cancer patients: Antibody subclasses and antigenic components. Gynecol. Oncol 2010, 116, 213–221. [Google Scholar]
- Koga, K.; Matsumoto, K.; Akiyoshi, T.; Kubo, M.; Yamanaka, N.; Tasaki, A.; Nakashima, H.; Nakamura, M.; Kuroki, S.; Tanaka, M.; Katano, M. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res 2005, 25, 3703–3707. [Google Scholar]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [Google Scholar]
- Taylor, D.D.; Zacharias, W.; Gercel-Taylor, C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol. Biol 2011, 728, 235–246. [Google Scholar]
- Simona, F.; Laura, S.; Simona, T.; Riccardo, A. Contribution of proteomics to understanding the role of tumor-derived exosomes in cancer progression: State of the art and new perspectives. Proteomics 2013, 13, 1581–1594. [Google Scholar] [Green Version]
- Eldh, M.; Lotvall, J.; Malmhall, C.; Ekstrom, K. Importance of RNA isolation methods for analysis of exosomal RNA: Evaluation of different methods. Mol. Immunol 2012, 50, 278–286. [Google Scholar]
- Lasser, C.; Eldh, M.; Lotvall, J. Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. 2012, e3037. [Google Scholar]
- Lasser, C. Identification and analysis of circulating exosomal microRNA in human body fluids. Methods Mol. Biol 2013, 1024, 109–128. [Google Scholar]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 2013, 14, 319. [Google Scholar]
- Nanosight. Available online: http://www.nanosight.com (on accessed 15 April 2013).
- Zheng, Y.; Campbell, E.C.; Lucocq, J.; Riches, A.; Powis, S.J. Monitoring the Rab27 associated exosome pathway using nanoparticle tracking analysis. Exp. Cell Res. 2012. [Google Scholar] [CrossRef]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar]
- Tian, T.; Wang, Y.; Wang, H.; Zhu, Z.; Xiao, Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J. Cell. Biochem 2010, 111, 488–496. [Google Scholar]
- Ristorcelli, E.; Beraud, E.; Verrando, P.; Villard, C.; Lafitte, D.; Sbarra, V.; Lombardo, D.; Verine, A. Human tumor nanoparticles induce apoptosis of pancreatic cancer cells. FASEB J 2008, 22, 3358–3369. [Google Scholar]
- Yang, Y.; Xiu, F.; Cai, Z.; Wang, J.; Wang, Q.; Fu, Y.; Cao, X. Increased induction of antitumor response by exosomes derived from interleukin-2 gene-modified tumor cells. J. Cancer Res. Clin. Oncol 2007, 133, 389–399. [Google Scholar]
- Dai, S.; Wei, D.; Wu, Z.; Zhou, X.; Wei, X.; Huang, H.; Li, G. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol. Ther 2008, 16, 782–790. [Google Scholar]
- Zhang, Y.; Wu, X.H.; Luo, C.L.; Zhang, J.M.; He, B.C.; Chen, G. Interleukin-12-anchored exosomes increase cytotoxicity of T lymphocytes by reversing the JAK/STAT pathway impaired by tumor-derived exosomes. Int. J. Mol. Med 2010, 25, 695–700. [Google Scholar]
- Zhang, Y.; Luo, C.L.; He, B.C.; Zhang, J.M.; Cheng, G.; Wu, X.H. Exosomes derived from IL-12-anchored renal cancer cells increase induction of specific antitumor response in vitro: A novel vaccine for renal cell carcinoma. Int. J. Oncol 2010, 36, 133–140. [Google Scholar]
- Chen, T.; Guo, J.; Yang, M.; Zhu, X.; Cao, X. Chemokine-containing exosomes are released from heat-stressed tumor cells via lipid raft-dependent pathway and act as efficient tumor vaccine. J. Immunol 2011, 186, 2219–2228. [Google Scholar]
- Peng, P.; Yan, Y.; Keng, S. Exosomes in the ascites of ovarian cancer patients: Origin and effects on anti-tumor immunity. Oncol. Rep 2011, 25, 749–762. [Google Scholar]
- Hakulinen, J.; Sankkila, L.; Sugiyama, N.; Lehti, K.; Keski-Oja, J. Secretion of active membrane type 1 matrix metalloproteinase (MMP-14) into extracellular space in microvesicular exosomes. J. Cell. Biochem 2008, 105, 1211–1218. [Google Scholar]
- McCready, J.; Sims, J.D.; Chan, D.; Jay, D.G. Secretion of extracellular hsp90alpha via exosomes increases cancer cell motility: A role for plasminogen activation. BMC Cancer 2010, 10, 294. [Google Scholar]
- Janowska-Wieczorek, A.; Wysoczynski, M.; Kijowski, J.; Marquez-Curtis, L.; Machalinski, B.; Ratajczak, J.; Ratajczak, M.Z. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int. J. Cancer 2005, 113, 752–760. [Google Scholar]
- Cho, J.A.; Park, H.; Lim, E.H.; Kim, K.H.; Choi, J.S.; Lee, J.H.; Shin, J.W.; Lee, K.W. Exosomes from ovarian cancer cells induce adipose tissue-derived mesenchymal stem cells to acquire the physical and functional characteristics of tumor-supporting myofibroblasts. Gynecol. Oncol 2011, 123, 379–386. [Google Scholar]
- Bobrie, A.; Krumeich, S.; Reyal, F.; Recchi, C.; Moita, L.F.; Seabra, M.C.; Ostrowski, M.; Thery, C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res 2012, 72, 4920–4930. [Google Scholar]
- Park, J.E.; Tan, H.S.; Datta, A.; Lai, R.C.; Zhang, H.; Meng, W.; Lim, S.K.; Sze, S.K. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteomics 2010, 9, 1085–1099. [Google Scholar]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringner, M.; Morgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. USA 2013. [Google Scholar] [CrossRef]
- Hong, B.S.; Cho, J.H.; Kim, H.; Choi, E.J.; Rho, S.; Kim, J.; Kim, J.H.; Choi, D.S.; Kim, Y.K.; Hwang, D.; et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics 2009, 10, 556. [Google Scholar]
- Ji, H.; Greening, D.W.; Barnes, T.W.; Lim, J.W.; Tauro, B.J.; Rai, A.; Xu, R.; Adda, C.; Mathivanan, S.; Zhao, W.; et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics 2013, 13, 1672–1686. [Google Scholar]
- Grange, C.; Tapparo, M.; Collino, F.; Vitillo, L.; Damasco, C.; Deregibus, M.C.; Tetta, C.; Bussolati, B.; Camussi, G. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res 2011, 71, 5346–5356. [Google Scholar]
- Zhu, W.; Huang, L.; Li, Y.; Zhang, X.; Gu, J.; Yan, Y.; Xu, X.; Wang, M.; Qian, H.; Xu, W. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett 2012, 315, 28–37. [Google Scholar]
- Mineo, M.; Garfield, S.H.; Taverna, S.; Flugy, A.; De Leo, G.; Alessandro, R.; Kohn, E.C. Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a Src-dependent fashion. Angiogenesis 2012, 15, 33–45. [Google Scholar]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med 2012, 18, 883–891. [Google Scholar] [Green Version]
- Baer, C.; Squadrito, M.L.; Iruela-Arispe, M.L.; De Palma, M. Reciprocal interactions between endothelial cells and macrophages in angiogenic vascular niches. Exp. Cell Res. 2013. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J 2002, 21, 4663–4670. [Google Scholar]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004, 23, 4051–4060. [Google Scholar]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res 2004, 32, 4776–4785. [Google Scholar]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar]
- Griffiths-Jones, S. The microRNA registry. Nucleic Acids Res 2004, 32, D109–D111. [Google Scholar]
- Lim, L.P.; Lau, N.C.; Garrett-Engele, P.; Grimson, A.; Schelter, J.M.; Castle, J.; Bartel, D.P.; Linsley, P.S.; Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 2005, 433, 769–773. [Google Scholar]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol 2010, 220, 126–139. [Google Scholar]
- Melo, S.A.; Esteller, M. Dysregulation of microRNAs in cancer: Playing with fire. FEBS Lett 2011, 585, 2087–2099. [Google Scholar]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. microRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar]
- Guo, Y.; Chen, Z.; Zhang, L.; Zhou, F.; Shi, S.; Feng, X.; Li, B.; Meng, X.; Ma, X.; Luo, M.; et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res 2008, 68, 26–33. [Google Scholar]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol 2009, 11, 1143–1149. [Google Scholar]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem 2010, 285, 17442–17452. [Google Scholar]
- Yao, B.; La, L.B.; Chen, Y.C.; Chang, L.J.; Chan, E.K. Defining a new role of GW182 in maintaining miRNA stability. EMBO Rep 2012, 13, 1102–1108. [Google Scholar]
- Ohshima, K.; Inoue, K.; Fujiwara, A.; Hatakeyama, K.; Kanto, K.; Watanabe, Y.; Muramatsu, K.; Fukuda, Y.; Ogura, S.; Yamaguchi, K.; et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One 2010, 5, e13247. [Google Scholar]
- Cifuentes, D.; Xue, H.; Taylor, D.W.; Patnode, H.; Mishima, Y.; Cheloufi, S.; Ma, E.; Mane, S.; Hannon, G.J.; Lawson, N.D.; et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010, 328, 1694–1698. [Google Scholar]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Stolz, D.B.; Sullivan, M.L.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar]
- Kogure, T.; Lin, W.L.; Yan, I.K.; Braconi, C.; Patel, T. Intercellular nanovesicle-mediated microRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 2011, 54, 1237–1248. [Google Scholar]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774. [Google Scholar]
- Takeshita, N.; Hoshino, I.; Mori, M.; Akutsu, Y.; Hanari, N.; Yoneyama, Y.; Ikeda, N.; Isozaki, Y.; Maruyama, T.; Akanuma, N.; et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br. J. Cancer 2013, 108, 644–652. [Google Scholar]
- Russo, F.; di Bella, S.; Nigita, G.; Macca, V.; Lagana, A.; Giugno, R.; Pulvirenti, A.; Ferro, A. miRandola: Extracellular circulating microRNAs database. PLoS One 2012, 7, e47786. [Google Scholar]
- Umezu, T.; Ohyashiki, K.; Kuroda, M.; Ohyashiki, J.H. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene 2012, 32, 2747–2755. [Google Scholar]
- Yang, M.; Chen, J.; Su, F.; Yu, B.; Su, F.; Lin, L.; Liu, Y.; Huang, J.D.; Song, E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol. Cancer 2011, 10, 117. [Google Scholar] [Green Version]
- Zhou, R.; O’Hara, S.P.; Chen, X.M. MicroRNA regulation of innate immune responses in epithelial cells. Cell. Mol. Immunol 2011, 8, 371–379. [Google Scholar]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar]
- Palma, J.; Yaddanapudi, S.C.; Pigati, L.; Havens, M.A.; Jeong, S.; Weiner, G.A.; Weimer, K.M.; Stern, B.; Hastings, M.L.; Duelli, D.M. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res 2012, 40, 9125–9138. [Google Scholar]
- Rana, S.; Malinowska, K.; Zoller, M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia 2013, 15, 281–295. [Google Scholar]
- Quesenberry, P.J.; Aliotta, J.M. Cellular phenotype switching and microvesicles. Adv. Drug Deliv. Rev 2010, 62, 1141–1148. [Google Scholar]
- Weigelt, B.; Bissell, M.J. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Semin. Cancer Biol 2008, 18, 311–321. [Google Scholar]
- Section on Breastfeeding. Breastfeeding and the use of human milk. Pediatrics 2012, 129, e827–e841.
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci 2012, 8, 118–123. [Google Scholar]
- Society, A.C. Breast Cancer Facts & Figures 2011–2012. Available online: http://www.cancer.org/research/cancerfactsfigures/breastcancerfactsfigures/breast-cancer-factsand-figures-2011-2012 (on accessed 15 April 2013).
- Locker, A.; Horrocks, C.; Gilmour, A.; Ellis, I.; Dowle, C.; Elston, C.; Blamey, R. Flow cytometric and histological analysis of ductal carcinoma in situ of the breast. Br. J. Surg 1990, 77, 564–567. [Google Scholar]
- Ottesen, G.; Christensen, I.; Larsen, J.; Christiansen, J.; Hansen, B.; Andersen, J. DNA analysis of in situ ductal carcinoma of the breast via flow cytometry. Cytometry 1995, 22, 168–176. [Google Scholar]
- Leal, C.; Schmitt, F.; Bento, M.; Maia, N.; Lopes, C. Ductal carcinoma in situ of the breast. Histologic categorization and its relationship to ploidy and immunohistochemical expression of hormone receptors, p53, and c-erbB-2 protein. Cancer 1995, 75, 2123–2131. [Google Scholar]
- Maguire, H.; Hellman, M.; Greene, M.; Yeh, I. Expression of c-erbB-2 in in situ and in adjacent invasive ductal adenocarcinomas of the female breast. Pathobiology 1992, 60, 117–121. [Google Scholar]
- Vos, C.; Ter Haar, N.; Peterse, J.; Cornelisse, C.; van de Vijver, M. Cyclin D1 gene amplification and overexpression are present in ductal carcinoma in situ of the breast. J. Pathol 1999, 187, 279–284. [Google Scholar]
- Radford, D.; Fair, K.; Phillips, N.; Ritter, J.; Steinbrueck, T.; Holt, M.; Donis-Keller, H. Allelotyping of ductal carcinoma in situ of the breast: Deletion of loci on 8p, 13q, 16q, 17p and 17q. Cancer Res 1995, 55, 3399–3405. [Google Scholar]
- Radford, D.; Phillips, N.; Fair, K.; Ritter, J.; Holt, M.; Donis-Keller, H. Allelic loss and the progression of breast cancer. Cancer Res 1995, 55, 5180–5183. [Google Scholar]
- Ma, X.J.; Dahiya, S.; Richardson, E.; Erlander, M.; Sgroi, D.C. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res 2009, 11, R7. [Google Scholar]
- Liu, C.; Yu, S.; Zinn, K.; Wang, J.; Zhang, L.; Jia, Y.; Kappes, J.C.; Barnes, S.; Kimberly, R.P.; Grizzle, W.E.; et al. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J. Immunol 2006, 176, 1375–1385. [Google Scholar]
- Higginbotham, J.N.; Demory Beckler, M.; Gephart, J.D.; Franklin, J.L.; Bogatcheva, G.; Kremers, G.J.; Piston, D.W.; Ayers, G.D.; McConnell, R.E.; Tyska, M.J.; et al. Amphiregulin exosomes increase cancer cell invasion. Curr. Biol 2011, 21, 779–786. [Google Scholar]
- O’Brien, K.; Rani, S.; Corcoran, C.; Wallace, R.; Hughes, L.; Friel, A.M.; McDonnell, S.; Crown, J.; Radomski, M.W.; O’Driscoll, L. Exosomes from triple-negative breast cancer cells can transfer phenotypic traits representing their cells of origin to secondary cells. Eur. J. Cancer 2013, 49, 1845–1859. [Google Scholar]
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012, 151, 1542–1556. [Google Scholar]
- Suetsugu, A.; Honma, K.; Saji, S.; Moriwaki, H.; Ochiya, T.; Hoffman, R.M. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv. Drug Deliv. Rev 2013, 65, 383–390. [Google Scholar]
- Sotiriou, C.; Pusztai, L. Gene-expression signatures in breast cancer. N. Engl. J. Med 2009, 360, 790–800. [Google Scholar]
- Blenkiron, C.; Goldstein, L.D.; Thorne, N.P.; Spiteri, I.; Chin, S.F.; Dunning, M.J.; Barbosa-Morais, N.L.; Teschendorff, A.E.; Green, A.R.; Ellis, I.O.; et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 2007, 8, R214. [Google Scholar]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar]
- Ciravolo, V.; Huber, V.; Ghedini, G.C.; Venturelli, E.; Bianchi, F.; Campiglio, M.; Morelli, D.; Villa, A.; Della Mina, P.; Menard, S.; et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J. Cell. Physiol 2012, 227, 658–667. [Google Scholar]
- Marleau, A.M.; Chen, C.S.; Joyce, J.A.; Tullis, R.H. Exosome removal as a therapeutic adjuvant in cancer. J. Transl. Med 2012, 10, 134. [Google Scholar]
- Safaei, R.; Larson, B.J.; Cheng, T.C.; Gibson, M.A.; Otani, S.; Naerdemann, W.; Howell, S.B. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol. Cancer Ther 2005, 4, 1595–1604. [Google Scholar]
- Ohno, S.; Takanashi, M.; Sudo, K.; Ueda, S.; Ishikawa, A.; Matsuyama, N.; Fujita, K.; Mizutani, T.; Ohgi, T.; Ochiya, T.; et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther 2013, 21, 185–191. [Google Scholar]
- Kooijmans, S.A.; Vader, P.; van Dommelen, S.M.; van Solinge, W.W.; Schiffelers, R.M. Exosome mimetics: A novel class of drug delivery systems. Int. J. Nanomedicine 2012, 7, 1525–1541. [Google Scholar]
| Cancer type | Clinical samples | Exosome isolation method | Major findings | Potential diagnostic microRNAs | Reference |
|---|---|---|---|---|---|
| Ovarian cancer | Sera from patients with serous papillary adenocarcinoma (n = 50); sera from age-matched controls with benign ovarian adenoma (n = 10); primary ovarian adenocarcinoma tumor cell cultures and matched patient sera (n = 6). | Magnetic activated cell sorting using beads coupled with anti-EpCAM. | Exosomal microRNA profiles were similar in ovarian cancer patient samples and distinctly different from benign disease samples. microRNAs were elevated in exosomes versus tumor cells (31 out of 467). | miR-21, miR-141, miR-200a, miR-200c, miR-203, miR-205 and miR-214 | [9] |
| Lung adenocarcinoma | Plasma from patients with lung adenocarcinoma (n = 27); plasma from controls (n = 9); matched plasma and lung tumor tissue (n = 4). | Size exclusion chromatography and magnetically activated cell sorting using beads coupled with anti-EpCAM. | No significant differences in exosome microRNA levels between microRNAs derived from circulating exosomes or from microRNAs from the primary tumor were observed. | miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212, miR-214 | [48] |
| Prostate cancer | Plasma from prostate cancer patients (n = 78); plasma from normal controls (n = 28); urine samples (n = 135); serum from patients with recurrent metastatic prostate cancer (n = 47) or non-recurrent disease (n = 72). | Filtration of plasma through a 1.2 μm filter, concentrated with a 150 kDa molecular weight cut-off. | The levels of 12 microRNAs were different between plasma exosomes of prostate cancer patients compared to control. Eleven microRNAs were present in significantly greater amounts in patients with metastases versus without. The association of exosomal miR-141 and miR-375 with metastases was confirmed in a second patient population. | miR-107, miR-130b, miR-141, miR-181a-2*, miR-2110, miR- 301a, miR-326, miR-331-3p, miR-432, miR-438, miR-574-3p, miR-625* | [110] |
| Esophageal squamous cell carcinoma | Serum from ESCC patients (n = 101); Serum from healthy controls (n = 46). | Sequential centrifugation, 0.22 μm filtration and ultracentrifugation. | miR-1246 was markedly elevated in serum and exosomes from ESCC patients and was a strong independent risk factor for poor survival. miR-1246 expression was not increased in ESCC tissue samples. | miR-1246 | [111] |
| Cell Line model | Major findings | Predominant microRNAs | Target genes or pathways | Reference |
|---|---|---|---|---|
| Human and mouse mast cells | Identified small RNAs, including 121 microRNAs and 1,300 specific mRNAs. Detected mouse exosomal RNA and new mouse proteins in human mast cells after treatment with mouse mast cell exosomes. Coined the term “exosomal shuttle RNA (esRNA)”. | let-7, miR-1, miR-15, miR-16, miR-181, miR-375. | None tested | [45] |
| Metastatic gastric cell line | Profiled microRNA expression by microarray in exosomes isolated from gastric cancer cells. let-7 microRNA family was enriched in exosomes. | let-7 family | None tested | [106] |
| Co-culture of IL-4-activated macrophages and breast cancer cells | miRNAs can be transferred from macrophages to breast cancer cells. miR-223 released by macrophages was found in MCF7 and MDA-MB-231 cells and promoted invasion. | miR-223 | Mef2c-β-catenin pathway | [114] |
| Mouse dendritic cells | Exosomal microRNA from dendritic cells can be transferred to a recipient dendritic cell and repress microRNA target mRNAs in the acceptor cell. | miR-148a, miR-451 | Luciferase reporter containing tandem microRNA target sequences | [108] |
| Leukemia cells and endothelial cells | Leukemia cells released microRNAs from the miR-17-92 cluster and were taken up by human umbilical vein endothelial cells (HUVECs) and repressed a target mRNA. Did not affect the growth of HUVEC cells, but did enhance cell migration and tube formation. | miR-92a | Integrin α5 | [113] |
| Hepatocellular carcinoma cells | Transmission of exosome microRNAs from hepatocellular carcinoma cells could contribute to the initiation and progression of hepatocellular carcinoma by targeting a tumor suppressor frequently lost in hepatocarcinogenesis. | miR-584, miR-517c, miR-378, miR-520f, miR-142-5p, miR-451, miR-518d, miR-215, miR-376a*, miR-133b, miR-367 | Transforming growth factor β activated kinase-1 (TAK1) pathway | [109] |
| Renal cancer stem cells | Microvesicles were secreted from human renal cell carcinoma that could trigger angiogenesis and premetastatic niche formation in a severe combined immunodeficient (SCID) mouse model. | miR-92, miR-141, miR-29a, miR-650, miR-151, miR-19b, miR-29c | Increase in VEGFR1 and MMP-9 expression | [87] |
| Breast cancer cells | Selective release of certain microRNA populations was demonstrated in malignant breast cancer cells that are retained in non-malignant mammary epithelial cells. | miR-451, miR-1246 | None tested | [49] |
| Metastatic rat adenocarcinoma cells | Exosomes were preferentially taken up by lymph node stroma cells and lung fibroblasts. The transferred microRNAs affected mRNA translation of many genes. | miR-494, miR-542-3p | Cadherin-17 and many proteases, adhesion molecules, chemokine ligands, cell cycle- and angiogenesis-promoting and oxidative stress response genes. | [118] |
| Lung cancer cell lines | miR-21 and miR-29a were secreted in exosomes and could bind to murine TLR7 and human TLR8 and trigger a Toll-like receptor (TLR)-mediated prometastatic inflammatory response that could lead to tumor growth and metastasis. | miR-21 and miR-29a | Toll-like receptor (TLR) 8 and 9 | [116] |
| Melanoma and normal melanocyte cells | The first in-depth screening to examine the entire exosome transcriptome, miRNome and proteome. Thousands of mRNAs and 15 microRNAs that are associated with melanoma progression and metastasis were identified. | let-7 family, miR-138, miR-125b, miR-130a, miR-34a, miR-196a, miR-199b-3p, miR-25, miR-27a, miR-200b, miR-23b, miR-146a, miR-613, miR-205, miR-149 | None tested | [51] |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hannafon, B.N.; Ding, W.-Q. Intercellular Communication by Exosome-Derived microRNAs in Cancer. Int. J. Mol. Sci. 2013, 14, 14240-14269. https://doi.org/10.3390/ijms140714240
Hannafon BN, Ding W-Q. Intercellular Communication by Exosome-Derived microRNAs in Cancer. International Journal of Molecular Sciences. 2013; 14(7):14240-14269. https://doi.org/10.3390/ijms140714240
Chicago/Turabian StyleHannafon, Bethany N., and Wei-Qun Ding. 2013. "Intercellular Communication by Exosome-Derived microRNAs in Cancer" International Journal of Molecular Sciences 14, no. 7: 14240-14269. https://doi.org/10.3390/ijms140714240




