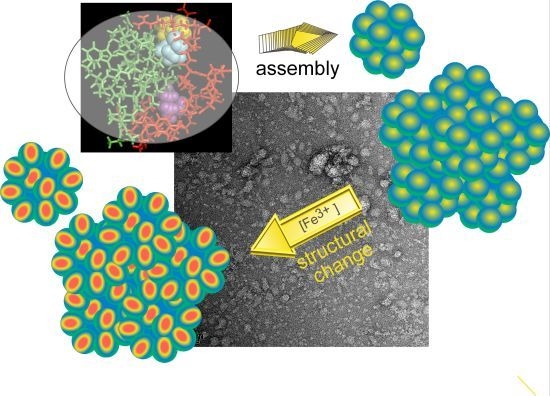
Integrated Self-Assembly of the Mms6 Magnetosome Protein to Form an Iron-Responsive Structure
Abstract
:1. Introduction
2. Results and Discussion
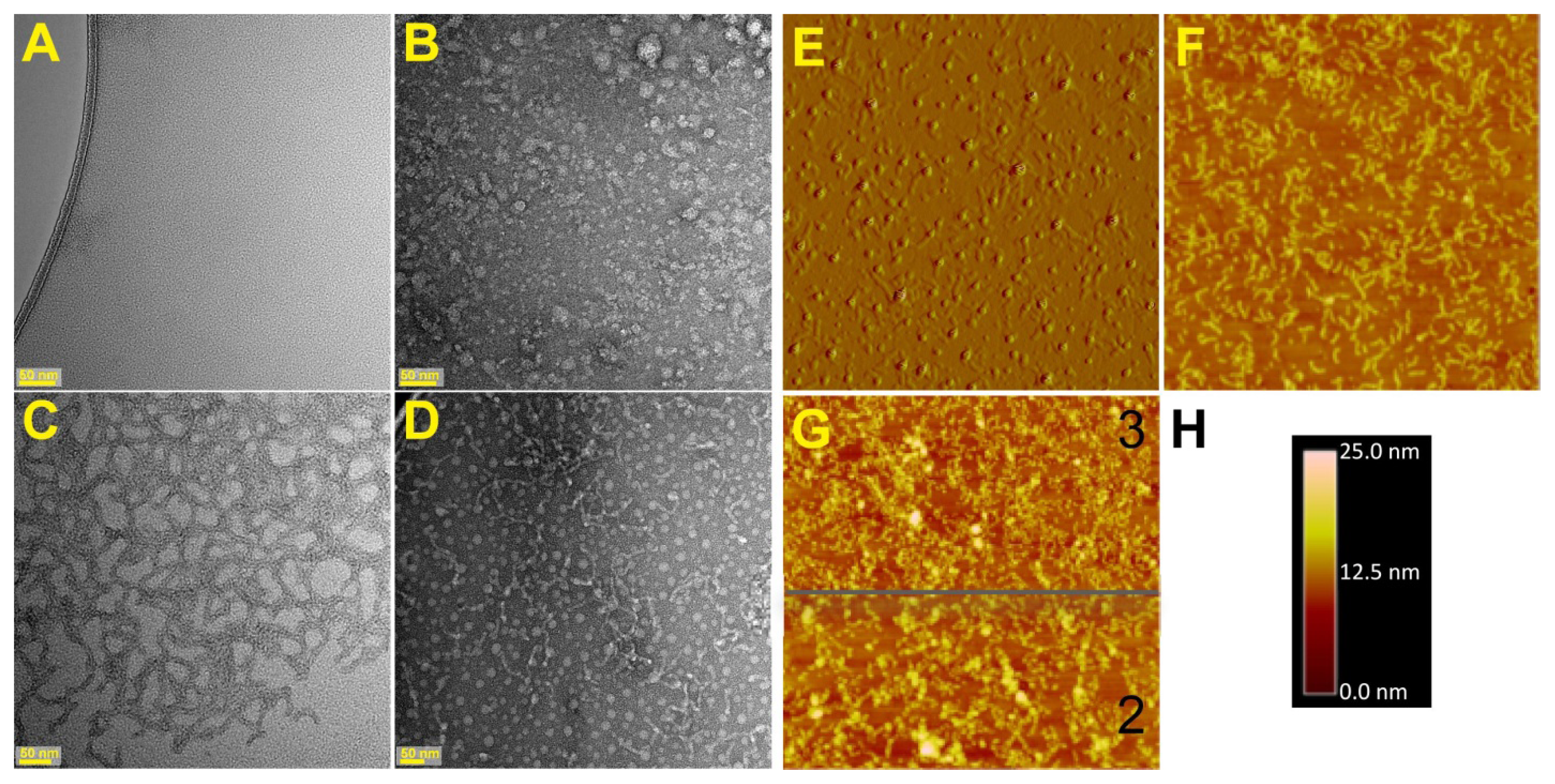
2.1. Visualization of the Self-Assembled Mms6
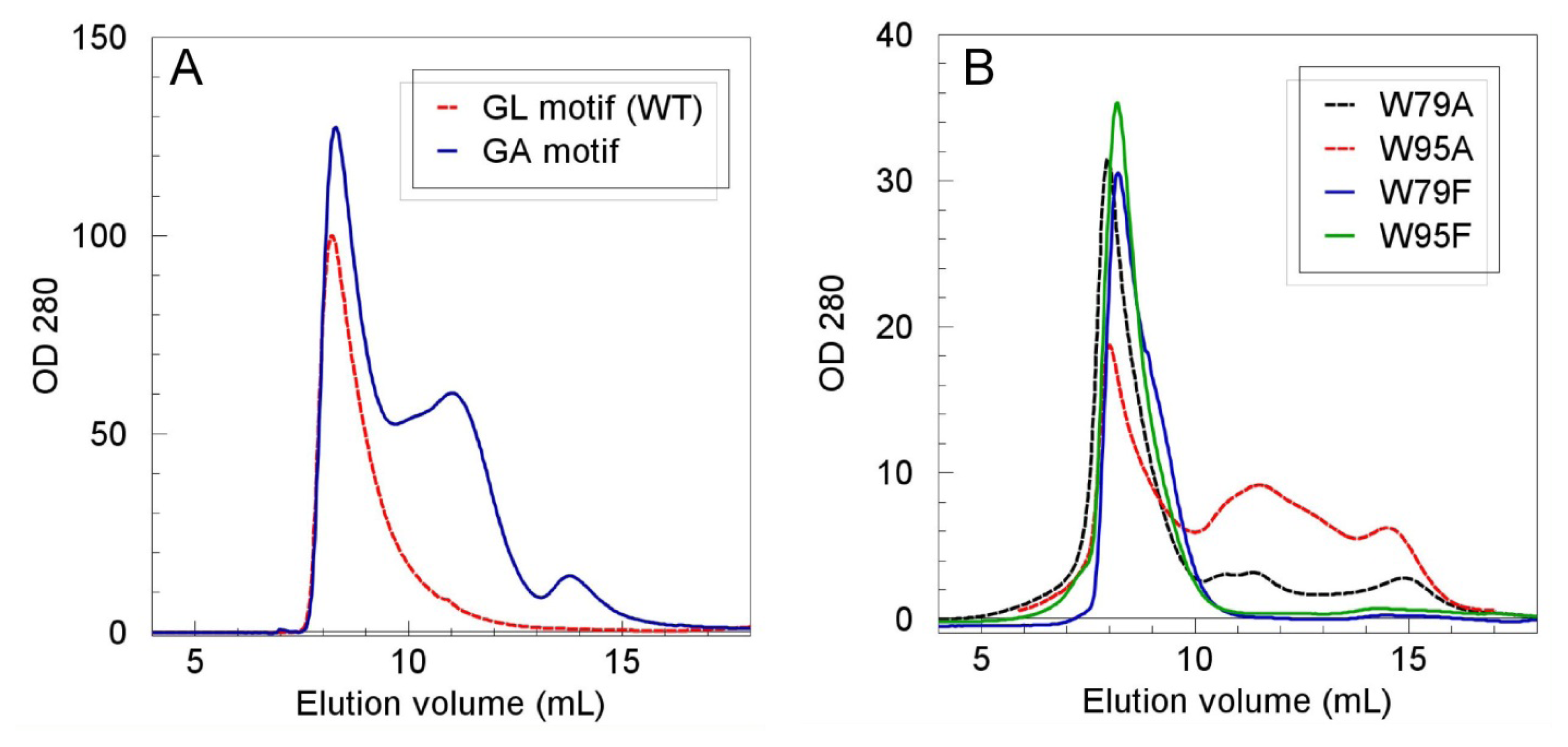
2.2. Role of the N-Terminal Domain in Promoting Mms6 Self-Assembly
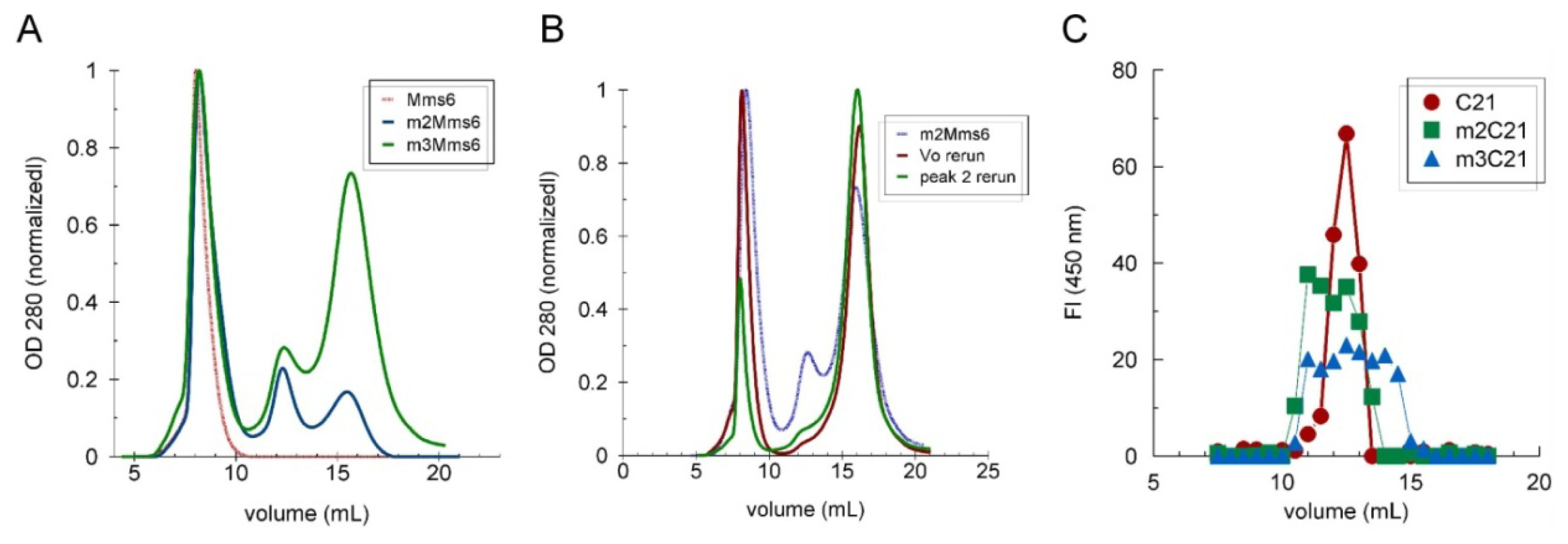
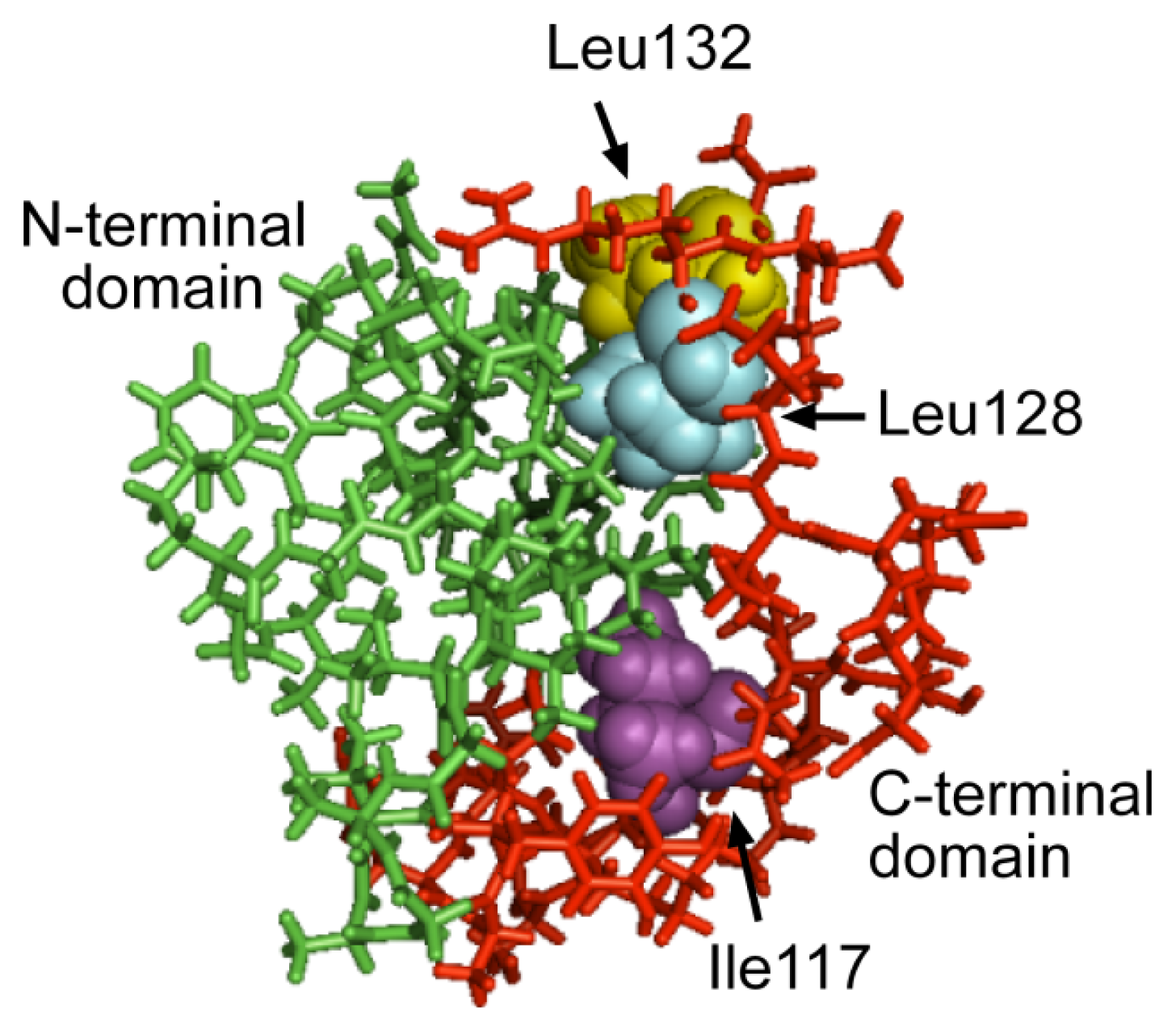
2.3. The C-Terminal Domain Contributes to the Stability of the Mms6 Micelles and Assembles in Multimeric Forms Independently of the N-Terminal Domain
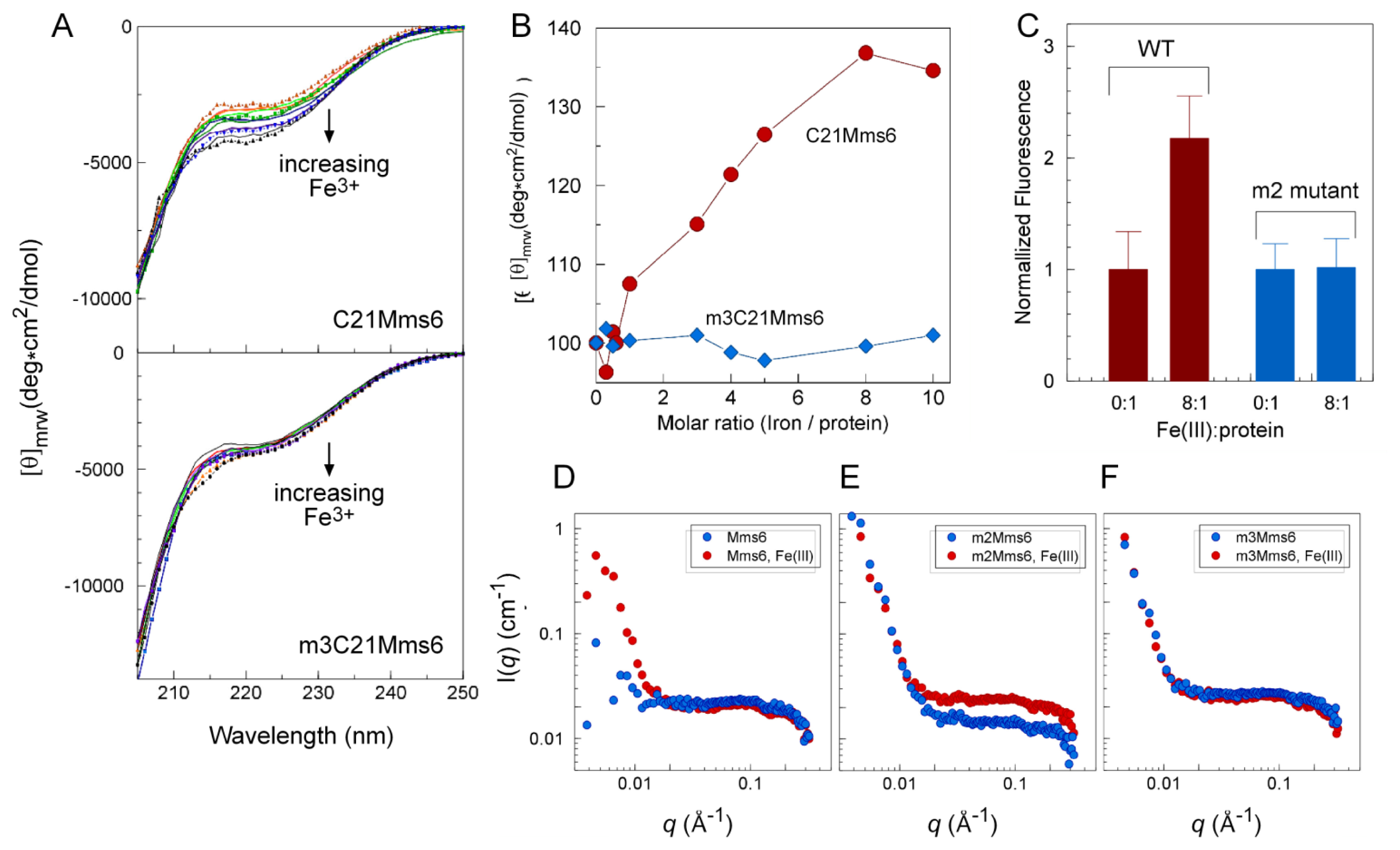
2.4. An Iron-Dependent Change in C-Terminal Domain Structure Is Transmitted to the N-terminal Domain of Mms6
3. Experimental Section
3.1. Protein Reagents and Preparation of Mutants
3.2. Atomic Force Microscopy (AFM)
3.3. Circular Dichroism (CD) Spectroscopy
3.4. Fluorescence Spectroscopy
3.5. Size Exclusion Chromatography
3.6. Small-Angle Neutron Scattering (SANS)
3.7. Transmission Electron Microscopy (TEM)
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Wang, L.; Nilsen-Hamilton, M. Biomineralization proteins: From vertebrates to bacteria. Front. Biol 2013, 8, 234–246. [Google Scholar]
- Evans, J.W.; Thiel, P.A. A little chemistry helps the big get bigger. Science 2010, 330, 599–600. [Google Scholar]
- Prozorov, T.; Palo, P.; Wang, L.; Nilsen-Hamilton, M.; Jones, D.; Orr, D.; Mallapragada, S.; Narasimhan, B. Cobalt ferrite nanocrystals: Out-performing magnetotactic bacteria. ACS Nano 2007, 1, 228–233. [Google Scholar]
- Prozorov, T.; Mallapragada, S.; Narasimhan, B.; Wang, L.; Palo, P.; Nilsen-Hamilton, M.; Williams, T.; Bazylinski, D.; Prozorov, R.; Canfield, P. Protein-mediated synthesis of uniform superparamagnetic magnetite nanocrystals. Adv. Funct. Mater 2007, 17, 951–957. [Google Scholar]
- Arakaki, A.; Webb, J.; Matsunaga, T. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem 2003, 278, 8745–8750. [Google Scholar]
- Tanaka, M.; Mazuyama, E.; Arakaki, A.; Matsunaga, T. Mms6 protein regulates crystal morphology during nano-sized magnetite biomineralization in vivo. J. Biol. Chem 2011, 286, 6386–6392. [Google Scholar]
- Wang, L.; Prozorov, T.; Palo, P.E.; Liu, X.; Vaknin, D.; Prozorov, R.; Mallapragada, S.; Nilsen-Hamilton, M. Self-assembly and biphasic iron-binding characteristics of Mms6, a bacterial protein that promotes the formation of superparamagnetic magnetite nanoparticles of uniform size and shape. Biomacromolecules 2012, 13, 98–105. [Google Scholar]
- Vargo, K.B.; Parthasarathy, R.; Hammer, D.A. Self-assembly of tunable protein suprastructures from recombinant oleosin. Proc. Natl. Acad. Sci. USA 2012, 109, 11657–11662. [Google Scholar]
- McDaniel, J.R.; Bhattacharyya, J.; Vargo, K.B.; Hassouneh, W.; Hammer, D.A.; Chilkoti, A. Self-assembly of thermally responsive nanoparticles of a genetically encoded peptide polymer by drug conjugation. Angew. Chem. Int. Ed 2013, 52, 1683–1687. [Google Scholar]
- Moitzi, C.; Portnaya, I.; Glatter, O.; Ramon, O.; Danino, D. Effect of temperature on self-assembly of bovine beta-casein above and below isoelectric pH. Structural analysis by cryogenic-transmission electron microscopy and small-angle X-ray scattering. Langmuir 2008, 24, 3020–3029. [Google Scholar]
- Portnaya, I.; Ben-Shoshan, E.; Cogan, U.; Khalfin, R.; Fass, D.; Ramon, O.; Danino, D. Self-assembly of bovine beta-casein below the isoelectric pH. J. Agric. Food Chem 2008, 56, 2192–2198. [Google Scholar]
- Wang, W.; Bu, W.; Wang, L.; Palo, P.E.; Mallapragada, S.; Nilsen-Hamilton, M.; Vaknin, D. Interfacial properties and iron binding to bacterial proteins that promote the growth of magnetite nanocrystals: X-ray reflectivity and surface spectroscopy studies. Langmuir 2012, 28, 4274–4282. [Google Scholar]
- Richter, M.; Kube, M.; Bazylinski, D.A.; Lombardot, T.; Glockner, F.O.; Reinhardt, R.; Schuler, D. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J. Bacteriol 2007, 189, 4899–4910. [Google Scholar]
- Mita, K.; Ichimura, S.; James, T.C. Highly repetitive structure and its organization of the silk fibroin gene. J. Mol. Evol 1994, 38, 583–592. [Google Scholar]
- Adamiano, A.; Bonacchi, S.; Calonghi, N.; Fabbri, D.; Falini, G.; Fermani, S.; Genovese, D.; Kralj, D.; Montalti, M.; NjegićDžakula, B.; et al. Structural changes in a protein fragment from abalone shell during the precipitation of calcium carbonate. Chem. Eur. J 2012, 18, 14367–14374. [Google Scholar]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008, 9, 40. [Google Scholar]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc 2010, 5, 725–738. [Google Scholar]
- Harris, J.R.; Plückthun, A.; Zahn, R. Transmission electron microscopy of GroEL, GroES, and the symmetrical GroEL/ES complex. J. Struct. Biol 1994, 112, 216–230. [Google Scholar]
| Mms6 protein | % protein in Vo |
|---|---|
| WT | 100 (15 repeats) |
| W79F | 96, 96 |
| W79A | 79, 83 |
| W95F | 97, 97 |
| W95A | 49, 70 |
| W79F,W95F | 75, 82 |
| L84A, L86A, L88A, L90A, L92A | 46, 54, 55 * |
| I117G | 94, 95 |
| L128G | 63, 86 |
| L132G | 29, 37 |
| Buffer content | # C21Mms6 units/multimer |
|---|---|
| water, pH 7.1 | >75 K (2) |
| 10 mM Pi, pH 7.5 | 7.3 ± 0.58 (3) |
| 10 mM Pi, 100 mM KCl, pH 7.5 | 4.0 (1) |
| 20 mM Tris, pH 7.5 | 2.0 ± 0.00 (3) |
| 20 mM Tris, 100 mM KCl, pH 7.5 | 4.1 ± 0.14 (2) |
| 20 mM Tris, 1.5 or 3 M KCl, pH 7.5 | 1.8 ± 0.38 (5) |
| 20 mM Tris, 6 M GnHCl, pH 7.5 | 1.1 (1) |
| 50 mM Formate or Citrate, pH 3 | 2.0 ± 0.08 (3) |
| 50 mM Formate or Citrate, pH 3 with FeCl3:protein = 8:1 | 2.1 ± 0.15 (4) |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Feng, S.; Wang, L.; Palo, P.; Liu, X.; Mallapragada, S.K.; Nilsen-Hamilton, M. Integrated Self-Assembly of the Mms6 Magnetosome Protein to Form an Iron-Responsive Structure. Int. J. Mol. Sci. 2013, 14, 14594-14606. https://doi.org/10.3390/ijms140714594
Feng S, Wang L, Palo P, Liu X, Mallapragada SK, Nilsen-Hamilton M. Integrated Self-Assembly of the Mms6 Magnetosome Protein to Form an Iron-Responsive Structure. International Journal of Molecular Sciences. 2013; 14(7):14594-14606. https://doi.org/10.3390/ijms140714594
Chicago/Turabian StyleFeng, Shuren, Lijun Wang, Pierre Palo, Xunpei Liu, Surya K. Mallapragada, and Marit Nilsen-Hamilton. 2013. "Integrated Self-Assembly of the Mms6 Magnetosome Protein to Form an Iron-Responsive Structure" International Journal of Molecular Sciences 14, no. 7: 14594-14606. https://doi.org/10.3390/ijms140714594