Identification and Expression Profile Analysis of Odorant Binding Proteins in the Oriental Fruit Fly Bactrocera dorsalis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of OBPs in B. dorsalis
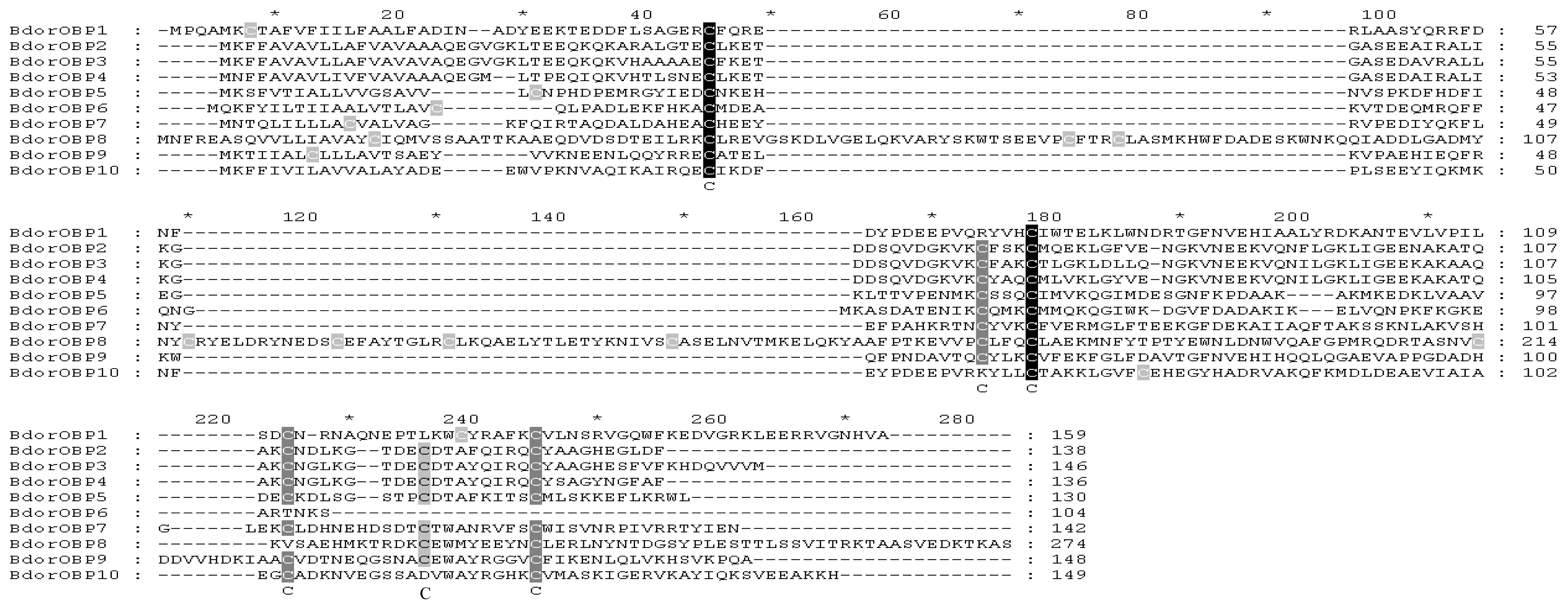
2.2. Homology Alignments of B. dorsalis OBPs
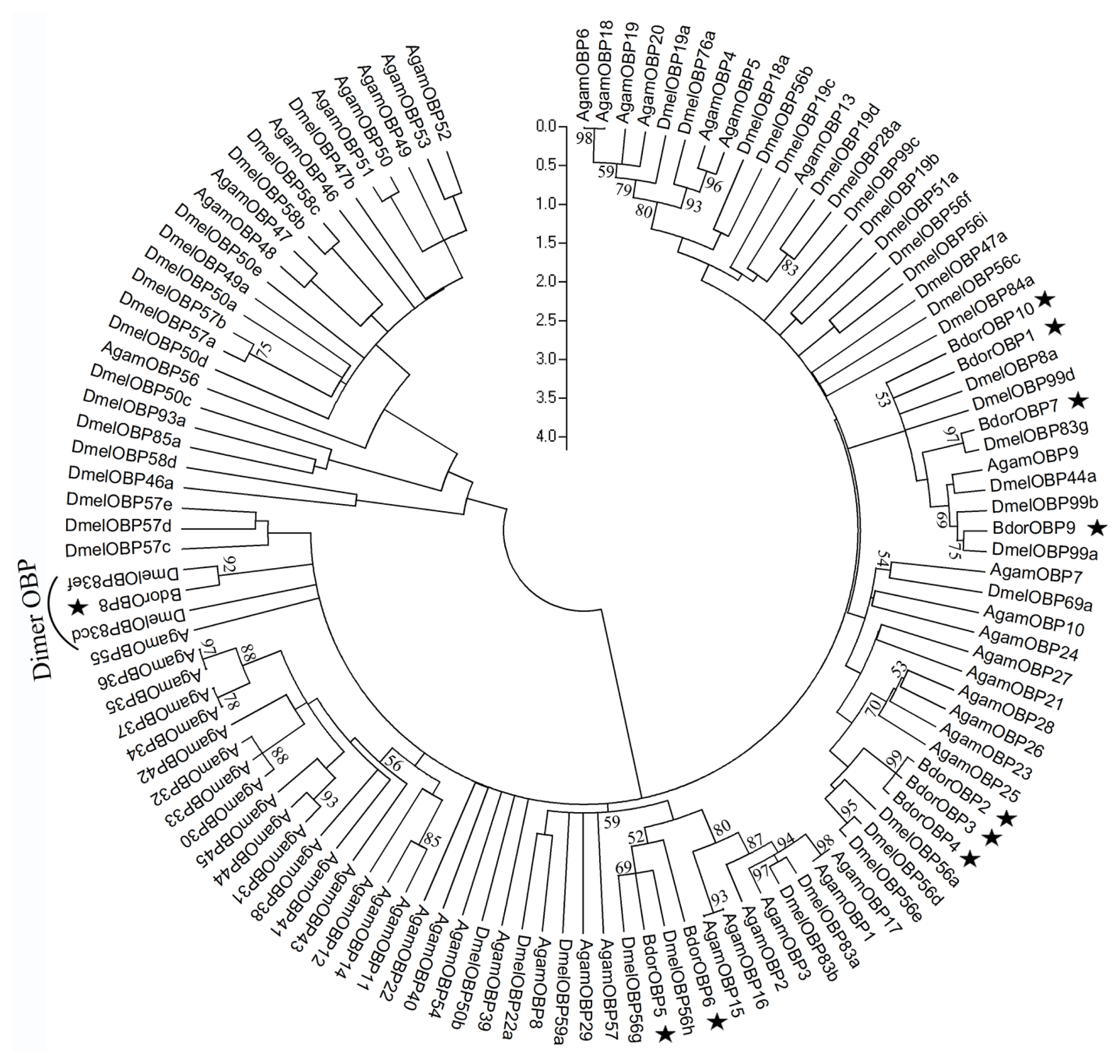
2.3. Phylogenetic Analysis of Dipteran OBPs
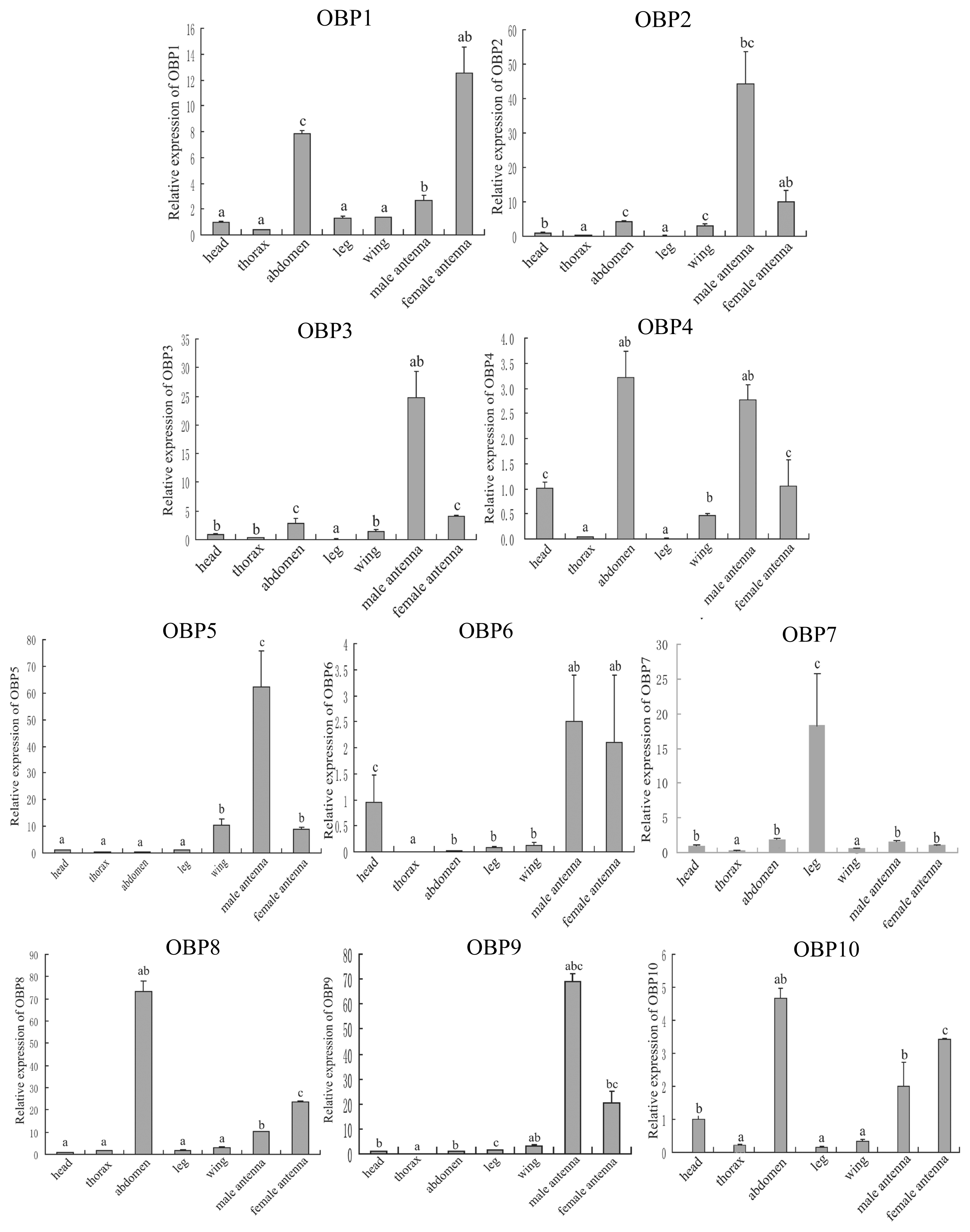
2.4. Tissue Expression Pattern of the OBP Genes
3. Experimental Section
3.1. Insect Rearing
3.2. Identification of the OBP Genes and Sequence Analysis
3.3. Phylogenetic Analysis
3.4. Total RNA Isolation and cDNA Synthesis
3.5. Expression Pattern Analysis of OBPs by Quantitative Real-time PCR
3.6. Statistical Analysis
4. Conclusions
Supplementary Information
ijms-14-14936-s001.pdfAcknowledgments
Conflict of Interest
References
- Justice, R.W.; Biessmann, H.; Walter, M.F.; Dimitratos, S.D.; Woods, D.F. Genomics spawns novel approaches to mosquito control. Bioessays 2003, 25, 1011–1020. [Google Scholar]
- Vogt, R.G.; Callahan, F.E.; Rogers, M.E.; Dickens, J.C. Odorant binding protein diversity and distribution among the insect orders, as indicated by LAP, an OBP-related protein of the true bug Lygus lineolaris (Hemiptera, Heteroptera). Chem. Senses 1999, 24, 481–495. [Google Scholar]
- Pelosi, P.; Maida, R. Odorant-binding proteins in insects. Comp. Biochem. Physiol. B Biochem. Mol. Biol 1995, 111, 503–514. [Google Scholar]
- Nakagawa, T.; Sakurai, T.; Nishioka, T.; Touhara, K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science 2005, 307, 1638–1642. [Google Scholar]
- Liu, R.; Lehane, S.; He, X.; Lehane, M.; Hertz-Fowler, C.; Berriman, M.; Pickett, J.A.; Field, L.M.; Zhou, J.J. Characterisations of odorant-binding proteins in the tsetse fly Glossina morsitans morsitans. Cell. Mol. Life Sci 2010, 67, 919–929. [Google Scholar]
- Xu, Y.L.; He, P.; Zhang, L.; Fang, S.Q.; Dong, S.L.; Zhang, Y.J.; Li, F. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genomics 2009, 10, 632. [Google Scholar]
- Zhou, J.J.; Huang, W.; Zhang, G.A.; Pickett, J.A.; Field, L.M. “Plus-C” odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae.”. Gene 2004, 327, 117–129. [Google Scholar]
- Zhou, J.J.; He, X.L.; Pickett, J.A.; Field, L.M. Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: Genome annotation and comparative analyses. Insect Mol. Biol 2008, 17, 147–163. [Google Scholar]
- Zhou, J.J.; Vieira, F.G.; He, X.L.; Smadja, C.; Liu, R.; Rozas, J.; Field, L.M. Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Mol. Biol 2010, 19, 113–122. [Google Scholar]
- Hekmat-Scafe, D.S.; Scafe, C.R.; McKinney, A.J.; Tanouye, M.A. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res 2002, 12, 1357–1369. [Google Scholar]
- Xu, P.X.; Zwiebel, L.J.; Smith, D.P. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol 2003, 12, 549–560. [Google Scholar]
- Wang, Y.; Wright, N.J.; Guo, H.; Xie, Z.; Svoboda, K.; Malinow, R.; Smith, D.P.; Zhong, Y. Genetic manipulation of the odor-evoked distributed neural activity in the Drosophila mushroom body. Neuron 2001, 29, 267–276. [Google Scholar]
- Kim, M.S.; Repp, A.; Smith, D.P. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics 1998, 150, 711–721. [Google Scholar]
- Xu, P.X.; Atkinson, R.; Jones, N.M.D.; Smith, D.P. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neurons 2005, 45, 193–200. [Google Scholar]
- Plettner, E.; Lazar, J.; Prestwich, E.G.; Prestwich, G.D. Discrimination of pheromone enantiomers by two pheromone binding proteins from the gypsy moth Lymantria dispar. Biochemistry 2000, 39, 8953–8962. [Google Scholar]
- Pelletier, J.; Guidolin, A.; Syed, Z.; Cornel, A.J.; Leal, W.S. Knockdown of a mosquito odorant-binding protein involved in the sensitive detection of oviposition attractants. J. Chem. Ecol 2010, 36, 245–248. [Google Scholar]
- Biessmann, H.; Andronopoulou, E.; Biessmann, M.R.; Douris, V.; Dimitratos, S.D.; Eliopoulos, E.; Guerin, P.M.; Iatrou, K.; Justice, R.W.; Krober, T.; et al. The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS One 2010, 5, e9471. [Google Scholar]
- Zhong, T.; Yin, J.; Deng, S.; Li, K.; Cao, Y. Fluorescence competition assay for the assessment of green leaf volatiles and trans-beta-farnesene bound to three odorant-binding proteins in the wheat aphid Sitobion avenae (Fabricius). J. Insect Physiol 2012, 58, 771–781. [Google Scholar]
- Prestwich, G.D.; Du, G.; LaForest, S. How is pheromone specificity encoded in proteins? Chem. Senses 1995, 20, 461–469. [Google Scholar]
- Maibeche-Coisne, M.; Jacquin-Joly, E.; Francois, M.C.; Nagnan-Le Meillour, P. Molecular cloning of two pheromone binding proteins in the cabbage armyworm Mamestra brassicae. Insect Biochem. Mol. Biol 1998, 28, 815–818. [Google Scholar]
- Mitaka, H.; Matsuo, T.; Miura, N.; Ishikawa, Y. Identification of odorant-binding protein genes from antennal expressed sequence tags of the onion fly, Delia antiqua. Mol. Biol. Rep 2011, 38, 1787–1792. [Google Scholar]
- Pelletier, J.; Leal, W.S. Genome analysis and expression patterns of odorant-binding proteins from the Southern House mosquito Culex pipiens quinquefasciatus. PLoS One 2009, 4, e6237. [Google Scholar]
- Xu, W.; Cornel, A.J.; Leal, W.S. Odorant-binding proteins of the malaria mosquito Anopheles funestus sensu stricto. PLoS One 2010, 5, e15403. [Google Scholar]
- Gong, D.P.; Zhang, H.J.; Zhao, P.; Xia, Q.Y.; Xiang, Z.H. The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics 2009, 10, 332. [Google Scholar]
- Grosse-Wilde, E.; Kuebler, L.S.; Bucks, S.; Vogel, H.; Wicher, D.; Hansson, B.S. Antennal transcriptome of Manduca sexta. Proc. Natl. Acad. Sci. USA 2011, 108, 7449–7454. [Google Scholar]
- Gotzek, D.; Robertson, H.M.; Wurm, Y.; Shoemaker, D. Odorant binding proteins of the red imported fire ant, Solenopsis invicta: An example of the problems facing the analysis of widely divergent proteins. PLoS One 2011, 6, e16289. [Google Scholar]
- Gu, S.H.; Wang, S.P.; Zhang, X.Y.; Wu, K.M.; Guo, Y.Y.; Zhou, J.J.; Zhang, Y.J. Identification and tissue distribution of odorant binding protein genes in the lucerne plant bug Adelphocoris lineolatus (Goeze). Insect Biochem. Mol. Biol 2011, 41, 254–263. [Google Scholar]
- Gomulski, L.M.; Dimopoulos, G.; Xi, Z.; Soares, M.B.; Bonaldo, M.F.; Malacrida, A.R.; Gasperi, G. Gene discovery in an invasive tephritid model pest species, the Mediterranean fruit fly, Ceratitis capitata. BMC Genomics 2008, 9, 243. [Google Scholar]
- Zheng, W.; Peng, T.; He, W.; Zhang, H. High-throughput sequencing to reveal genes involved in reproduction and development in Bactrocera dorsalis (Diptera: Tephritidae). PLoS One 2012, 7, e36463. [Google Scholar]
- Vieira, F.G.; Sanchez-Gracia, A.; Rozas, J. Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: Purifying selection and birth-and-death evolution. Genome Biol. 2007, 8. [Google Scholar] [CrossRef]
- Shanbhag, S.R.; Muller, B.; Steinbrecht, R.A. Atlas of olfactory organs of Drosophila melanogaster 2. Internal organization and cellular architecture of olfactory sensilla. Arthropod Struct. Dev 2000, 29, 211–229. [Google Scholar]
- Sun, Y.L.; Huang, L.Q.; Pelosi, P.; Wang, C.Z. Expression in antennae and reproductive organs suggests a dual role of an odorant-binding protein in two sibling Helicoverpa species. PLoS One 2012, 7, e30040. [Google Scholar]
- Li, X.; Zhang, M.; Zhang, H. RNA interference of four genes in adult Bactrocera dorsalis by feeding their dsRNAs. PLoS One 2011, 6, e17788. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 2012, 40, W597–W603. [Google Scholar]
- Oliver, T.; Schmidt, B.; Nathan, D.; Clemens, R.; Maskell, D. Using reconfigurable hardware to accelerate multiple sequence alignment with ClustalW. Bioinformatics 2005, 21, 3431–3432. [Google Scholar]
- Nicholas, K.B.; Nicholas, H.B., Jr; Deerfield, D.W., II. GeneDoc: Analysis and visualization of genetic variation. Embnew. News 1997, 4, 14. [Google Scholar]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res 2012, 40, D302–D305. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 2011, 39, W475–W478. [Google Scholar]
- Zheng, W.; Zhu, C.; Peng, T.; Zhang, H. Odorant receptor co-receptor Orco is upregulated by methyl eugenol in male Bactrocera dorsalis (Diptera: Tephritidae). J. Insect Physiol 2012, 58, 1122–1127. [Google Scholar]
- Shen, G.M.; Dou, W.; Niu, J.Z.; Jiang, H.B.; Yang, W.J.; Jia, F.X.; Hu, F.; Cong, L.; Wang, J.J. Transcriptome analysis of the oriental fruit fly (Bactrocera dorsalis). PLoS One 2011, 6, e29127. [Google Scholar]

| Gene name | Accession number | Length | Drosophila gene | Subject ID | E-value | Identity (%) |
|---|---|---|---|---|---|---|
| BdorOBP1 | KC559112 | 159 | OBP8a | ACY92783 | 1 × 10−14 | 33 |
| BdorOBP2 | KC559113 | 138 | OBP56d | NP_611444 | 2 × 10−21 | 45 |
| BdorOBP3 | KC559114 | 146 | OBP56d | NP_611444 | 8 × 10−22 | 47 |
| BdorOBP4 | KC559115 | 136 | OBP56d | NP_611444 | 1 ×10−19 | 44 |
| BdorOBP5 | KC559116 | 130 | OBP56g | NP_611447 | 3 ×10−14 | 34 |
| BdorOBP6 | KC559117 | 104 * | OBP56h | ABW78079 | 1 × 10−11 | 36 |
| BdorOBP7 | KC559118 | 142 | OBP83g | AAN13232 | 2 × 10−54 | 63 |
| BdorOBP8 | KC559119 | 274 | OBP83ef | AAF51918 | 1 × 10−69 | 47 |
| BdorOBP9 | KC559120 | 148 | OBP99a | ACT22143 | 2 × 10−56 | 56 |
| BdorOBP10 | KC559121 | 149 | OBP99c | ACT22281 | 3 × 10−46 | 54 |
| Primer name | (5′→3′) Nucleotide sequence |
|---|---|
| BdorOBP1RTF | GGCGAGCGTTGTTTCCAG |
| BdorOBP1RTR | ATCGGCACCAGCACTTCC |
| BdorOBP2RTF | TTCTTCGCTGTTGCTGTTTTGC |
| BdorOBP2RTR | AGAAGCATTTGACTTTGCCATC |
| BdorOBP3RTF | TTGACCGAGGAGCAGAAAC |
| BdorOBP3RTR | TTGACTTTGCCATCGACTT |
| BdorOBP4RTF | GAACTTCTTCGCTGTTGCTG |
| BdorOBP4RTR | CTTGACTGTCATCGCCTTTG |
| BdorOBP5RTF | CCACGACTTCATTGAGGGTA |
| BdorOBP5RTR | CCACAGCAGCAACTAATTTATC |
| BdorOBP6RTF | ACGAAGCCAAAGTCACGG |
| BdorOBP6RTR | GCATCAAAGACGCCATCC |
| BdorOBP7RTF | GTTGCAGGCAAATTCCAGAT |
| BdorOBP7RTR | AGAGTCCCATTCGCTCCACA |
| BdorOBP8RTF | CGAAGTCGGCAGCAAAGA |
| BdorOBP8RTR | TCGTCCGCATCGAACCAG |
| BdorOBP9RTF | GCGATGCCGACCATGATGAC |
| BdorOBP9RTR | ACCACCACGATAAGCCCACT |
| BdorOBP10RTF | GTGTCTTCTGCGAACATGAGG |
| BdorOBP10RTR | CACTTGTGACCACGATAGGC |
| β-actinRTF | CTCGTCCAACCGTTCATACC |
| β-actinRTR | CTGACCTGCCCACTGAAGTT |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zheng, W.; Peng, W.; Zhu, C.; Zhang, Q.; Saccone, G.; Zhang, H. Identification and Expression Profile Analysis of Odorant Binding Proteins in the Oriental Fruit Fly Bactrocera dorsalis. Int. J. Mol. Sci. 2013, 14, 14936-14949. https://doi.org/10.3390/ijms140714936
Zheng W, Peng W, Zhu C, Zhang Q, Saccone G, Zhang H. Identification and Expression Profile Analysis of Odorant Binding Proteins in the Oriental Fruit Fly Bactrocera dorsalis. International Journal of Molecular Sciences. 2013; 14(7):14936-14949. https://doi.org/10.3390/ijms140714936
Chicago/Turabian StyleZheng, Weiwei, Wei Peng, Chipan Zhu, Qun Zhang, Giuseppe Saccone, and Hongyu Zhang. 2013. "Identification and Expression Profile Analysis of Odorant Binding Proteins in the Oriental Fruit Fly Bactrocera dorsalis" International Journal of Molecular Sciences 14, no. 7: 14936-14949. https://doi.org/10.3390/ijms140714936




