PCA3 and PCA3-Based Nomograms Improve Diagnostic Accuracy in Patients Undergoing First Prostate Biopsy
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characteristics of Our Validation Cohort
2.2. Diagnostic Performance of the Urinary PCA3 Test
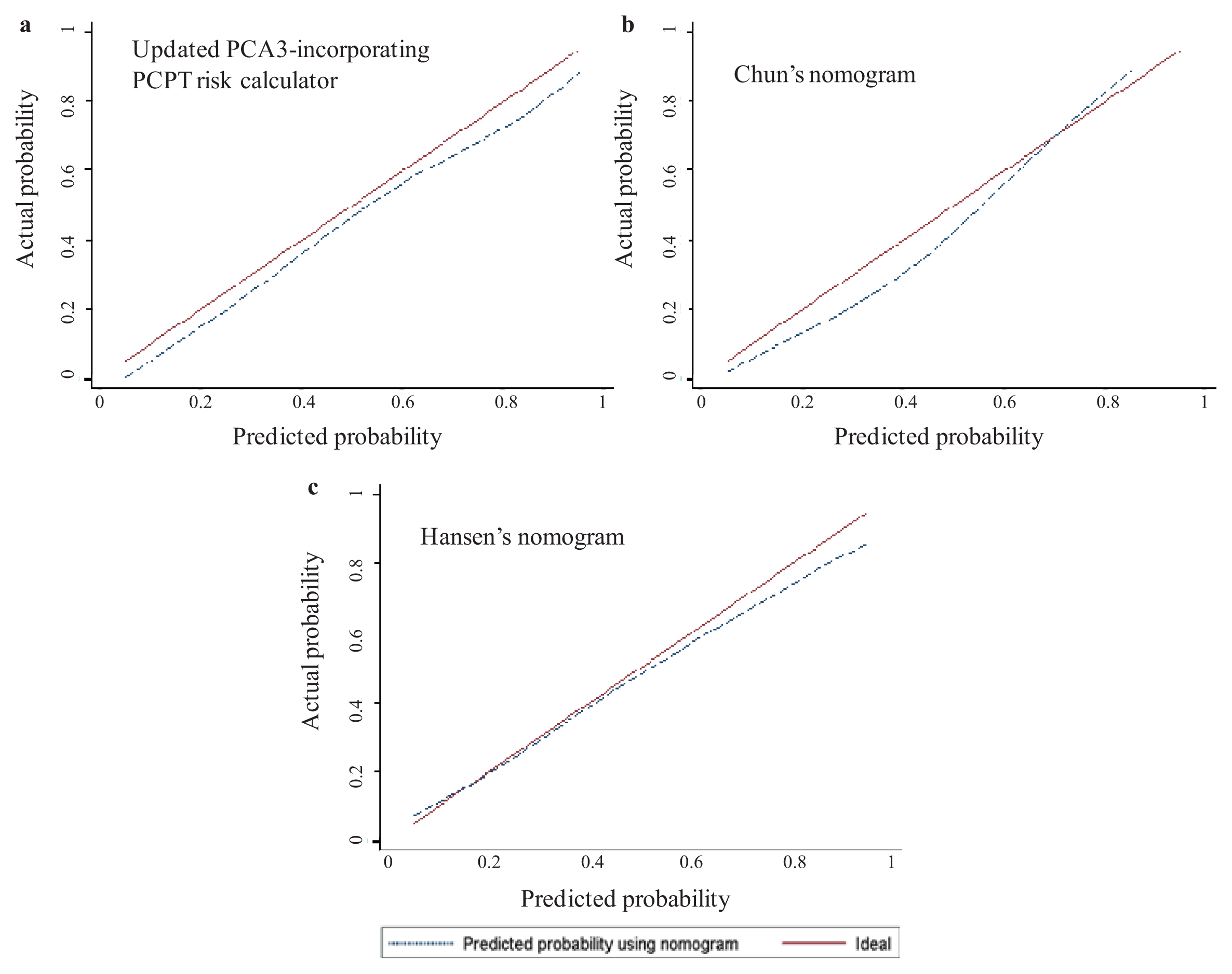
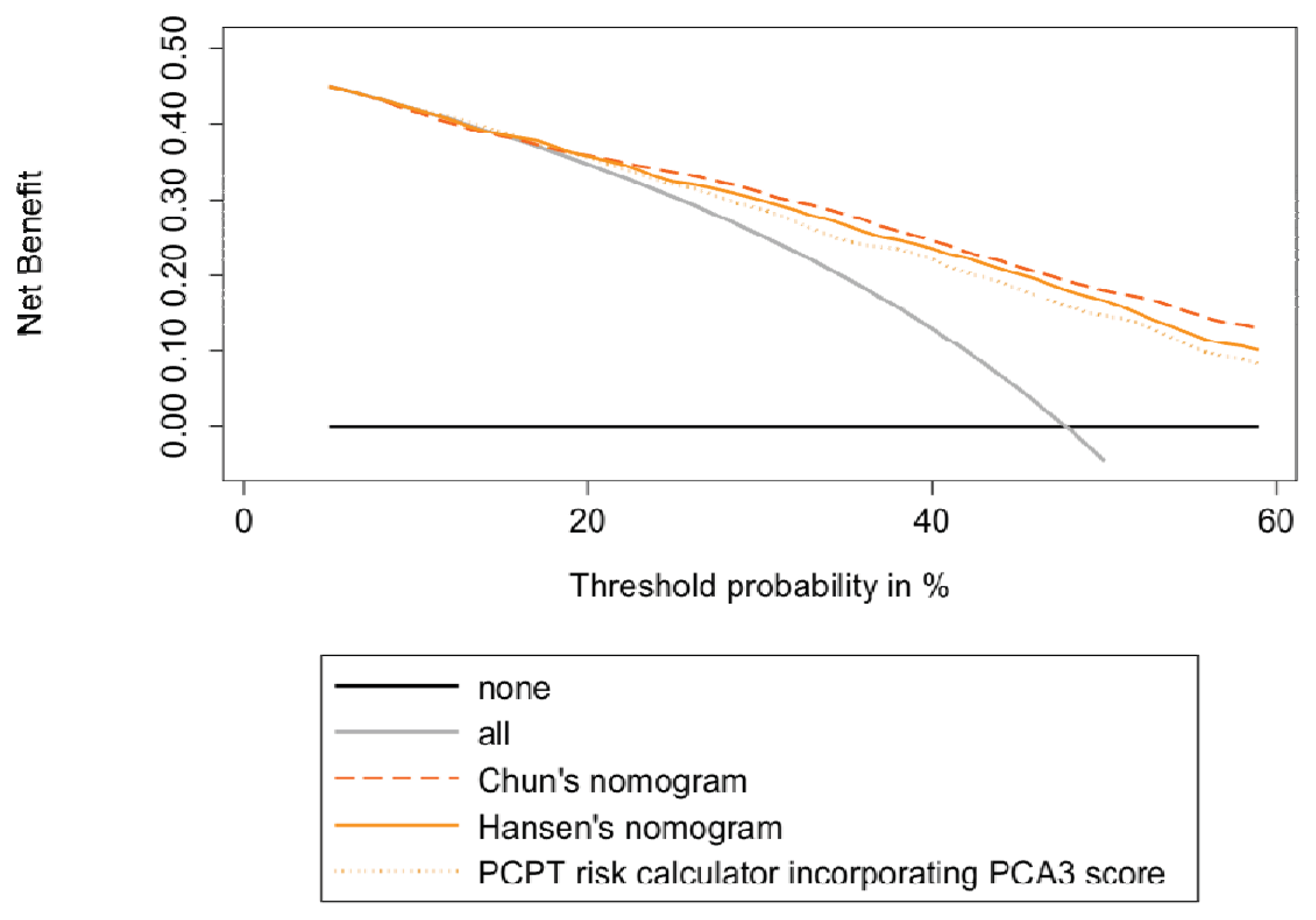
2.3. Head-to-Head Comparisons of the 3 Available Urinary PCA3-Based Nomograms
2.4. Discussion
3. Experimental Section
3.1. Patients and Study Design
3.2. Biochemical Assays and Nomograms
3.3. Prostate Biopsies
3.4. Statistics
4. Conclusions
Supplementary Information
ijms-14-17767-s001.pdfAcknowledgments
Conflicts of Interest
References
- Wilt, T.J.; Ahmed, H.U. Prostate cancer screening and the management of clinically localized disease. BMJ 2013, 346, f325. [Google Scholar]
- Hessels, D.; Klein Gunnewiek, J.M.; van Oort, I.; Karthaus, H.F.; van Leenders, G.J.; van Balken, B.; Kiemeney, L.A.; Witjes, J.A.; Schalken, J.A. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol 2003, 44, 8–15. [Google Scholar]
- Auprich, M.; Bjartell, A.; Chun, F.K.; de la Taille, A.; Freedland, S.J.; Haese, A.; Schalken, J.; Stenzl, A.; Tombal, B.; van der Poel, H. Contemporary role of prostate cancer antigen 3 in the management of prostate cancer. Eur. Urol 2011, 60, 1045–1054. [Google Scholar]
- Vlaeminck-Guillem, V.; Ruffion, A.; Andre, J.; Devonec, M.; Paparel, P. Urinary prostate cancer 3 test: Toward the age of reason? Urology 2010, 75, 447–453. [Google Scholar]
- Marks, L.S.; Bostwick, D.G. Prostate cancer specificity of pca3 gene testing: Examples from clinical practice. Rev. Urol 2008, 10, 175–181. [Google Scholar]
- Roobol, M.J.; Schroder, F.H.; van Leeuwen, P.; Wolters, T.; van den Bergh, R.C.; van Leenders, G.J.; Hessels, D. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: Exploring the value of PCA3 for a first-line diagnostic test. Eur. Urol 2010, 58, 475–481. [Google Scholar]
- Goode, R.R.; Marshall, S.J.; Duff, M.; Chevli, E.; Chevli, K.K. Use of PCA3 in detecting prostate cancer in initial and repeat prostate biopsy patients. Prostate 2013, 73, 48–53. [Google Scholar]
- Bollito, E.; de Luca, S.; Cicilano, M.; Passera, R.; Grande, S.; Maccagnano, C.; Cappia, S.; Milillo, A.; Montorsi, F.; Scarpa, R.M.; et al. Prostate cancer gene 3 urine assay cutoff in diagnosis of prostate cancer: A validation study on an Italian patient population undergoing first and repeat biopsy. Anal. Quant. Cytol. Histol 2012, 34, 96–104. [Google Scholar]
- Deras, I.L.; Aubin, S.M.; Blase, A.; Day, J.R.; Koo, S.; Partin, A.W.; Ellis, W.J.; Marks, L.S.; Fradet, Y.; Rittenhouse, H.; et al. PCA3: A molecular urine assay for predicting prostate biopsy outcome. J. Urol 2008, 179, 1587–1592. [Google Scholar]
- Ramos, C.G.; Valdevenito, R.; Vergara, I.; Anabalon, P.; Sanchez, C.; Fulla, J. PCA3 sensitivity and specificity for prostate cancer detection in patients with abnormal PSA and/or suspicious digital rectal examination. First Latin American experience. Urol. Oncol. 2012. [Google Scholar] [CrossRef]
- Schilling, D.; Hennenlotter, J.; Munz, M.; Bokeler, U.; Sievert, K.D.; Stenzl, A. Interpretation of the prostate cancer gene 3 in reference to the individual clinical background: Implications for daily practice. Urol. Int 2010, 85, 159–165. [Google Scholar]
- De la Taille, A.; Irani, J.; Graefen, M.; Chun, F.; de Reijke, T.; Kil, P.; Gontero, P.; Mottaz, A.; Haese, A. Clinical evaluation of the PCA3 assay in guiding initial biopsy decisions. J. Urol 2011, 185, 2119–2125. [Google Scholar]
- Ferro, M.; Bruzzese, D.; Perdona, S.; Mazzarella, C.; Marino, A.; Sorrentino, A.; di Carlo, A.; Autorino, R.; di Lorenzo, G.; Buonerba, C.; et al. Predicting prostate biopsy outcome: Prostate health index (phi) and prostate cancer antigen 3 (PCA3) are useful biomarkers. Clin. Chim. Acta 2012, 413, 1274–1278. [Google Scholar]
- Hansen, J.; Auprich, M.; Ahyai, S.A.; de la Taille, A.; van Poppel, H.; Marberger, M.; Stenzl, A.; Mulders, P.F.; Huland, H.; Fisch, M.; et al. Initial prostate biopsy: Development and internal validation of a biopsy-specific nomogram based on the prostate cancer antigen 3 assay. Eur. Urol 2013, 63, 201–209. [Google Scholar]
- Perdona, S.; Bruzzese, D.; Ferro, M.; Autorino, R.; Marino, A.; Mazzarella, C.; Perruolo, G.; Longo, M.; Spinelli, R.; di Lorenzo, G.; et al. Prostate health index (phi) and prostate cancer antigen 3 (PCA3) significantly improve diagnostic accuracy in patients undergoing prostate biopsy. Prostate 2013, 73, 227–235. [Google Scholar]
- Crawford, E.D.; Rove, K.O.; Trabulsi, E.J.; Qian, J.; Drewnowska, K.P.; Kaminetsky, J.C.; Huisman, T.K.; Bilowus, M.L.; Freedman, S.J.; Glover, W.L., Jr; et al. Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: A prospective study of 1,962 cases. J. Urol. 2012, 188, 1726–1731. [Google Scholar]
- Haese, A.; de la Taille, A.; van Poppel, H.; Marberger, M.; Stenzl, A.; Mulders, P.F.; Huland, H.; Abbou, C.C.; Remzi, M.; Tinzl, M.; et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. Eur. Urol 2008, 54, 1081–1088. [Google Scholar]
- Ochiai, A.; Okihara, K.; Kamoi, K.; Iwata, T.; Kawauchi, A.; Miki, T.; Fors, Z. Prostate cancer gene 3 urine assay for prostate cancer in Japanese men undergoing prostate biopsy. Int. J. Urol 2011, 18, 200–205. [Google Scholar]
- Salagierski, M.; Mulders, P.; Schalken, J.A. Predicting prostate biopsy outcome using a PCA3-based nomogram in a Polish cohort. Anticancer Res 2013, 33, 553–557. [Google Scholar]
- Ankerst, D.P.; Groskopf, J.; Day, J.R.; Blase, A.; Rittenhouse, H.; Pollock, B.H.; Tangen, C.; Parekh, D.; Leach, R.J.; Thompson, I. Predicting prostate cancer risk through incorporation of prostate cancer gene 3. J. Urol 2008, 180, 1303–1308. [Google Scholar]
- Chun, F.K.; de la Taille, A.; van Poppel, H.; Marberger, M.; Stenzl, A.; Mulders, P.F.; Huland, H.; Abbou, C.C.; Stillebroer, A.B.; van Gils, M.P.; et al. Prostate cancer gene 3 (PCA3): Development and internal validation of a novel biopsy nomogram. Eur. Urol 2009, 56, 659–668. [Google Scholar]
- Auprich, M.; Haese, A.; Walz, J.; Pummer, K.; de la Taille, A.; Graefen, M.; de Reijke, T.; Fisch, M.; Kil, P.; Gontero, P.; et al. External validation of urinary PCA3-Based nomograms to individually predict prostate biopsy outcome. Eur. Urol 2010, 8, 727–732. [Google Scholar]
- Wu, A.K.; Reese, A.C.; Cooperberg, M.R.; Sadetsky, N.; Shinohara, K. Utility of PCA3 in patients undergoing repeat biopsy for prostate cancer. Prostate Cancer Prostatic Dis 2012, 15, 100–105. [Google Scholar]
- Epstein, J.I.; Walsh, P.C.; Carmichael, M.; Brendler, C.B. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. JAMA 1994, 271, 368–374. [Google Scholar]
- Leyten, G.H.; Hessels, D.; Jannink, S.A.; Smit, F.P.; de Jong, H.; Cornel, E.B.; de Reijke, T.M.; Vergunst, H.; Kil, P.; Knipscheer, B.C.; et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur. Urol. 2013. [Google Scholar] [CrossRef]
- Abern, M.R.; Freedland, S.J. Prostate cancer antigen 3 to select men for prostate biopsy: Stop, go, or proceed with caution? Eur. Urol 2013, 63, 210–211. [Google Scholar]
- Perdona, S.; Cavadas, V.; di Lorenzo, G.; Damiano, R.; Chiappetta, G.; del Prete, P.; Franco, R.; Azzarito, G.; Scala, S.; Arra, C.; et al. Prostate cancer detection in the “grey area” of prostate-specific antigen below 10 ng/mL: Head-to-head comparison of the updated PCPT calculator and chun’s nomogram, two risk estimators incorporating prostate cancer antigen 3. Eur. Urol 2011, 59, 81–87. [Google Scholar]
- Dong, F.; Jones, J.S.; Stephenson, A.J.; Magi-Galluzzi, C.; Reuther, A.M.; Klein, E.A. Prostate cancer volume at biopsy predicts clinically significant upgrading. J. Urol 2008, 179, 896–900. [Google Scholar]
- Vlaeminck-Guillem, V.; Devonec, M.; Colombel, M.; Rodriguez-Lafrasse, C.; Decaussin-Petrucci, M.; Ruffion, A. Urinary PCA3 score predicts prostate cancer multifocality. J. Urol 2011, 185, 1234–1239. [Google Scholar]
- Groskopf, J.; Aubin, S.M.; Deras, I.L.; Blase, A.; Bodrug, S.; Clark, C.; Brentano, S.; Mathis, J.; Pham, J.; Meyer, T.; et al. APTIMA PCA3 molecular urine test: Development of a method to aid in the diagnosis of prostate cancer. Clin. Chem 2006, 52, 1089–1095. [Google Scholar]
- Vickers, A.J.; Elkin, E.B. Decision curve analysis: A novel method for evaluating prediction models. Med. Decis. Making 2006, 26, 565–574. [Google Scholar]
| Entire initial biopsy cohort | No cancer at initial biopsy | LGPCa at initial biopsy | HGPCa at initial biopsy | p-Value * | |
|---|---|---|---|---|---|
| No. of patients (%) | 594 (100) | 318 (54) | 148 (25) | 128 (22) | - |
| Age, year | <0.0001 | ||||
| Median | 63 | 62 | 64 | 65 | |
| IQR | 58–67 | 58–66 | 59–68 | 61–70 | |
| DRE | <0.001 | ||||
| Unsuspicious, no. (%) | 519 (87) | 293 (92) | 136 (92) | 90 (70) | |
| Suspicious, no. (%) | 75 (13) | 25 (8) | 12 (8) | 38 (30) | |
| Familial history of PCa | 0.428 | ||||
| No (%) | 505 (85) | 273 (86) | 121 (82) | 111 (87) | |
| Yes (%) | 89 (15) | 45 (14) | 27 (18) | 17 (13) | |
| Prostate volume *, mL | <0.0001 | ||||
| Median | 39.5 | 42 | 37 | 30 | |
| IQR | 28–50 | 32–55 | 29–48 | 24–47 | |
| Serum PSA, ng/mL | 0.012 | ||||
| Median | 5.9 | 6 | 5.7 | 6.1 | |
| IQR | 4.7–7.9 | 4.6–7.6 | 4.6–7.4 | 5.1–8.9 | |
| ≥2.5 ng/mL | 579 (97) | 308 (97) | 143 (97) | 128 (100) | 0.119 |
| ≥4 ng/mL | 525 (88) | 279 (88) | 133 (90) | 113 (88) | 0.799 |
| ≥10 ng/mL | 70 (12) | 32 (10) | 17 (11) | 21 (16) | 0.170 |
| Urinary PCA3 score | <0.0001 | ||||
| Median | 30 | 18 | 48 | 52 | |
| IQR | 15–65 | 10–42 | 25–94 | 26–100 | |
| ≥35 | 265 (45) | 90 (28) | 93 (63) | 82 (64) | <0.001 |
| <35 | 329 (55) | 228 (72) | 55 (37) | 46 (36) | |
| >21 | 364 (61) | 138 (43) | 117 (79) | 109 (85) | <0.001 |
| ≤21 | 230 (39) | 180 (57) | 31 (21) | 19 (15) |
| Multivariate analysis | ||||||||
|---|---|---|---|---|---|---|---|---|
| Base model | Base model + continuous PCA3 score | Base model + PCA3 cutoff of 21 | Base model + PCA3 cutoff of 35 | |||||
| OR (95% CI) | p-Value | OR (95% CI) | p-Value | OR (95% CI) | p-Value | OR (95% CI) | p-Value | |
| Age, year | 1.08 (1.05–1.11) | <0.001 | 1.05 (1.02–1.09) | <0.001 | 1.06 (1.02–1.09) | <0.001 | 1.05 (1.02–1.08) | 0.001 |
| DRE | 1.09 (1.03–1.15) | 0.004 | 1.08 (1.02–1.15) | 0.006 | 1.08 (1.02–1.15) | 0.008 | 1.09 (1.03–1.16) | 0.004 |
| Prostate volume, cm3 | 0.96 (0.95–0.97) | <0.001 | 0.96 (0.95–0.97) | <0.001 | 0.96 (0.95–0.98) | <0.001 | 0.96 (0.95–0.97) | <0.001 |
| Serum PSA, ng/mL | 1.10 (1.03–1.17) | 0.004 | 1.10 (1.02–1.18) | 0.008 | 1.08 (1.01–1.16) | 0.027 | 1.09 (1.02–1.17) | 0.015 |
| Urinary PCA3 score | - | - | 1.01 (1.01–1.01) | <0.001 | 5.00 (3.36–7.45) | <0.001 | 4.21 (2.88–6.15) | <0.001 |
| AUC | 0.714 | 0.780 | 0.781 | 0.780 | ||||
| IC 95% | (0.672–0.755) | (0.743–0.818) | (0.744–0.818) | (0.742–0.817) | ||||
| p-Value * | - | p < 0.0001 | p < 0.0001 | p < 0.0001 | ||||
| PA | 66% | 72% | 71% | 73% | ||||
| IC 95% | (62%–70%) | (68%–76%) | (68%–75%) | (69%–76%) | ||||
| Increment in PA * | - | +6% | +5% | +7% | ||||
| p-Value * | - | p = 0.033 | p = 0.060 | p = 0.017 | ||||
| Nomogram | Probability cutoff (%) | Biopsies performed | Biopsies not performed a | Biopsies not performed in men without PCa b | Any PCa detected c | Any PCa missed | NPV for PCa prediction | HGPCa detected | HGPCa missed | NPV for HGPCa prediction |
|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | % | n (%) | n (%) | % | ||
| None | 536 (100) | 0 (0) | 0 (0) | 256 (100) | 0 (0) | 100 | 122 (100) | 0 (0) | 100 | |
| PCPT [20] | 10 | 531 (99) | 5 (1) | 5 (2) | 256 (100) | 0 (0) | 100 | 122 (100) | 0 (0) | 100 |
| 20 | 512 (96) | 24 (4) | 22 (8) | 254 (99) | 2 (1) | 92 | 121 (99) | 1 (1) * | 98 | |
| 30 | 462 (86) | 74 (14) | 66 (22) | 244 (95) | 12 (5) | 84 | 118 (97) | 4 (3) * | 95 | |
| 40 | 376 (70) | 160 (30) | 123 (44) | 219 (86) | 37 (14) | 77 | 108 (89) | 14 (11) ** | 93 | |
| 50 | 275 (51) | 261 (49) | 184 (66) | 179 (70) | 77 (30) | 71 | 90 (74) | 32 (26) *** | 88 | |
| Chun [21] | 10 | 516 (96) | 20 (4) | 17 (6) | 253 (99) | 3 (1) | 85 | 122 (100) | 0 (0) | 100 |
| 20 | 462 (86) | 74 (14) | 63 (23) | 245 (96) | 11 (4) | 85 | 121 (99) | 1 (1) * | 98 | |
| 30 | 375 (70) | 161 (30) | 134 (48) | 229 (89) | 27 (11) | 83 | 115 (94) | 7 (6) * | 96 | |
| 40 | 342 (64) | 194 (36) | 154 (55) | 216 (84) | 40 (16) | 79 | 108 (89) | 14 (11) * | 93 | |
| 50 | 249 (46) | 287 (54) | 204 (73) | 173 (68) | 83 (32) | 71 | 93 (76) | 29 (24) **** | 90 | |
| Hansen [14] | 10 | 524 (98) | 12 (2) | 11 (4) | 255 (99.6) | 1 (0.4) | 92 | 122 (100) | 0 (0) | 100 |
| 20 | 466 (87) | 70 (13) | 66 (22) | 248 (97) | 8 (3) | 89 | 121 (99) | 1 (1) * | 98 | |
| 30 | 390 (73) | 146 (27) | 119 (43) | 229 (89) | 27 (11) | 82 | 115 (94) | 7 (6) * | 96 | |
| 40 | 345 (64) | 191 (36) | 149 (53) | 214 (84) | 42 (16) | 78 | 108 (89) | 14 (11) * | 93 | |
| 50 | 292 (54) | 244 (46) | 179 (64) | 191 (75) | 65 (25) | 73 | 97 (80) | 25 (20) ** | 90 | |
| Race a | Age (year) | Serum PSA (ng/mL) | Family history (yes/no) | DRE e | Negative previous biopsy(ies) (yes/no) | Prostate volume (cm3) | Urinary PCA3 score f | |
|---|---|---|---|---|---|---|---|---|
| PCPT [20] | + | + b | + | + | + | + | + | |
| Chun [21] | + | + c | + | + | + | + | ||
| Hansen [14] | + | + d | + | + | + |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ruffion, A.; Devonec, M.; Champetier, D.; Decaussin-Petrucci, M.; Rodriguez-Lafrasse, C.; Paparel, P.; Perrin, P.; Vlaeminck-Guillem, V. PCA3 and PCA3-Based Nomograms Improve Diagnostic Accuracy in Patients Undergoing First Prostate Biopsy. Int. J. Mol. Sci. 2013, 14, 17767-17780. https://doi.org/10.3390/ijms140917767
Ruffion A, Devonec M, Champetier D, Decaussin-Petrucci M, Rodriguez-Lafrasse C, Paparel P, Perrin P, Vlaeminck-Guillem V. PCA3 and PCA3-Based Nomograms Improve Diagnostic Accuracy in Patients Undergoing First Prostate Biopsy. International Journal of Molecular Sciences. 2013; 14(9):17767-17780. https://doi.org/10.3390/ijms140917767
Chicago/Turabian StyleRuffion, Alain, Marian Devonec, Denis Champetier, Myriam Decaussin-Petrucci, Claire Rodriguez-Lafrasse, Philippe Paparel, Paul Perrin, and Virginie Vlaeminck-Guillem. 2013. "PCA3 and PCA3-Based Nomograms Improve Diagnostic Accuracy in Patients Undergoing First Prostate Biopsy" International Journal of Molecular Sciences 14, no. 9: 17767-17780. https://doi.org/10.3390/ijms140917767




