Functional Diversity of Genes for the Biosynthesis of Paeoniflorin and Its Derivatives in Paeonia
Abstract
:1. Introduction
2. Results
2.1. Identification of 24 Paeoniflorin and Gallic Acid Biosynthesis-Related Genes in P. lactiflora and Other Related Plants
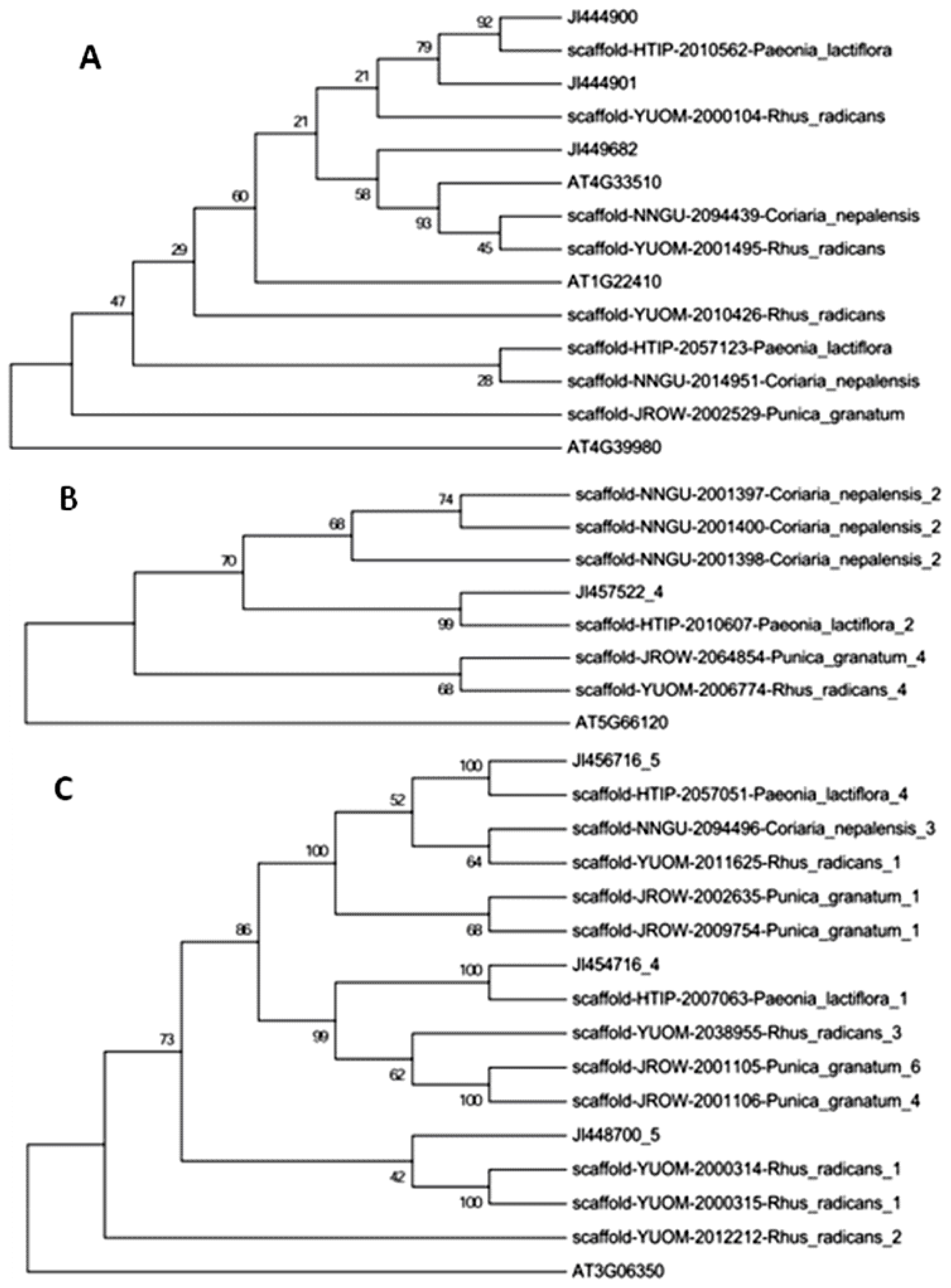
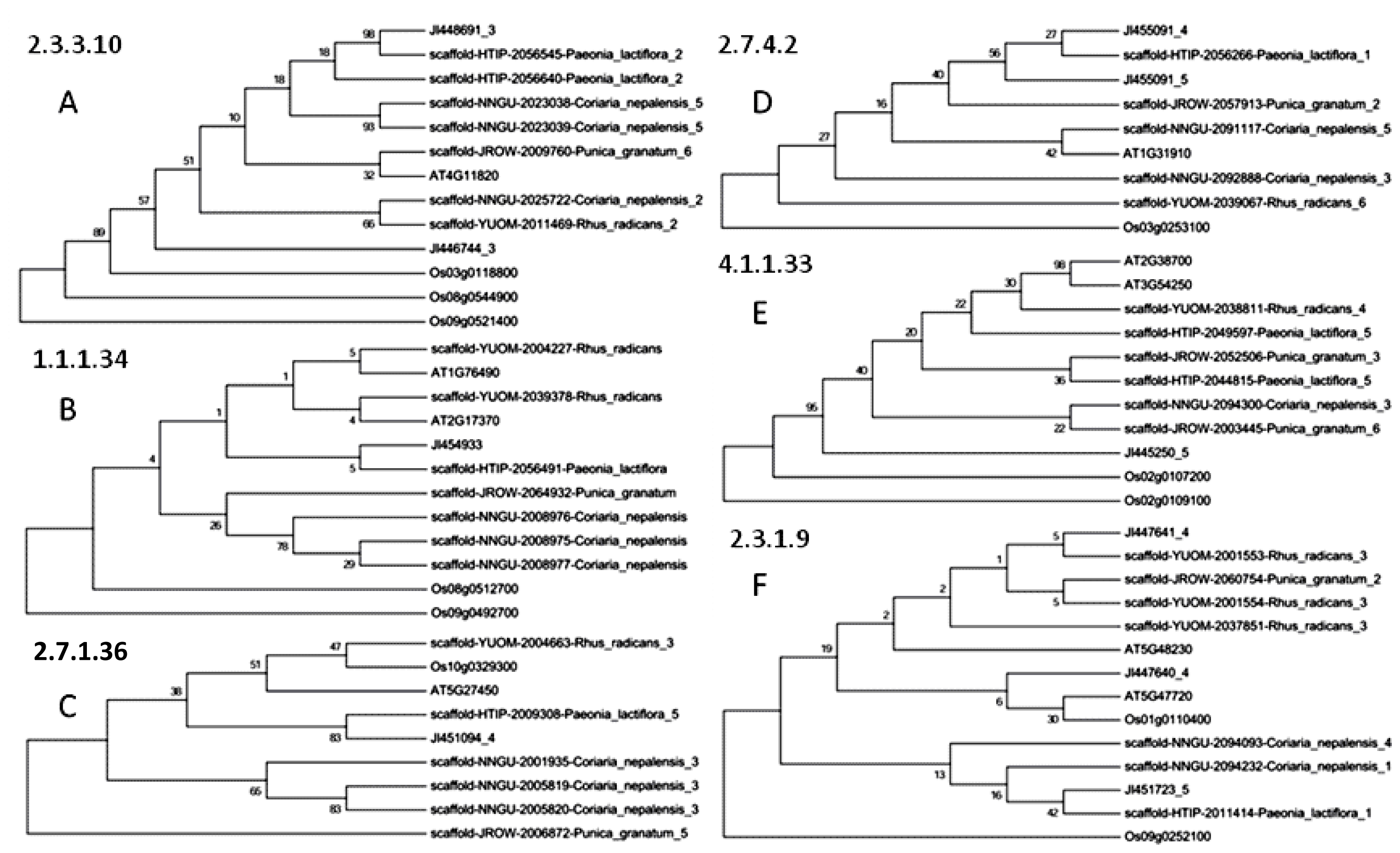
2.2. Phylogenetic Analysis of Paeoniflorin and Gallic Acid Biosynthesis-Related Orthologs
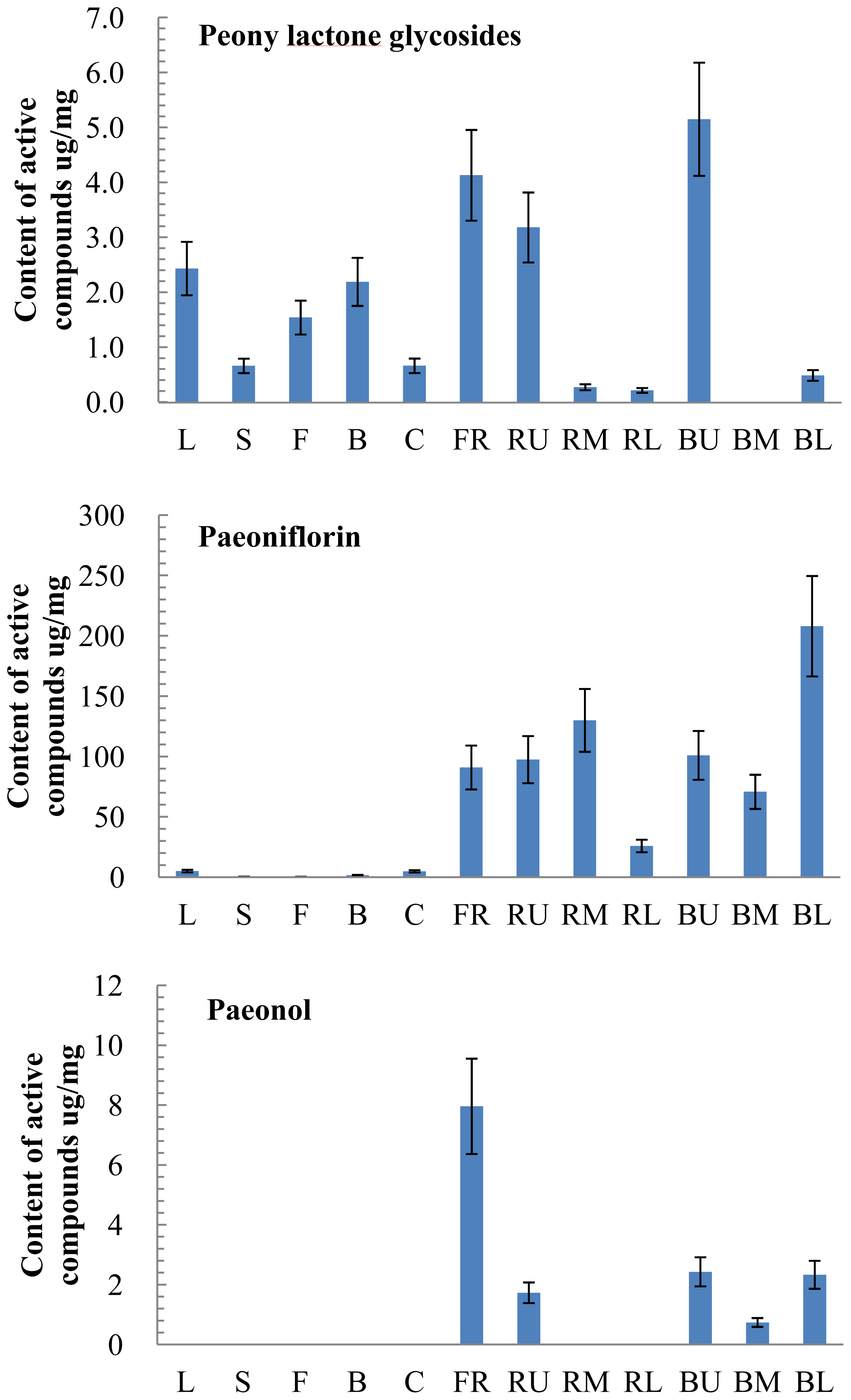
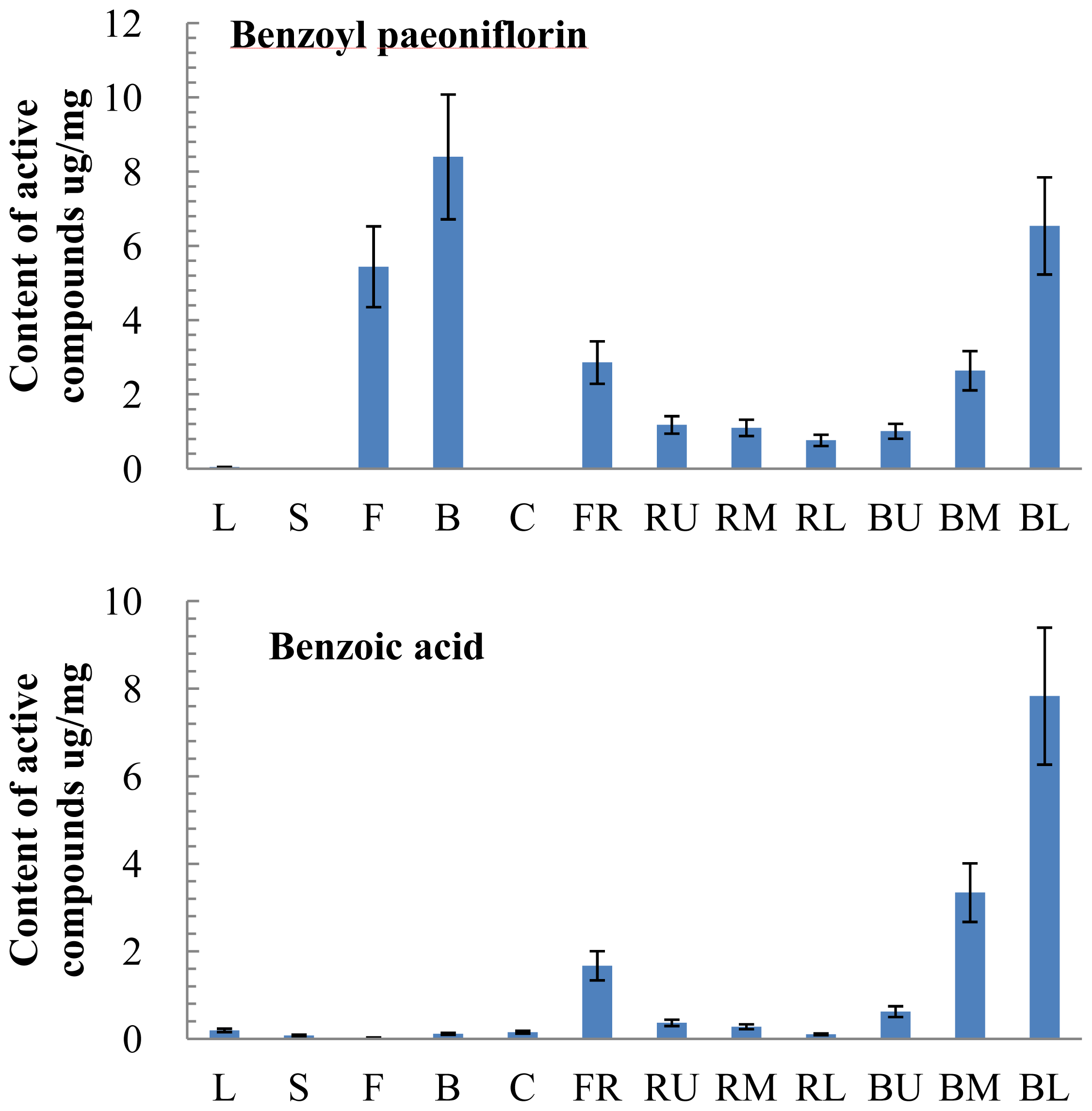
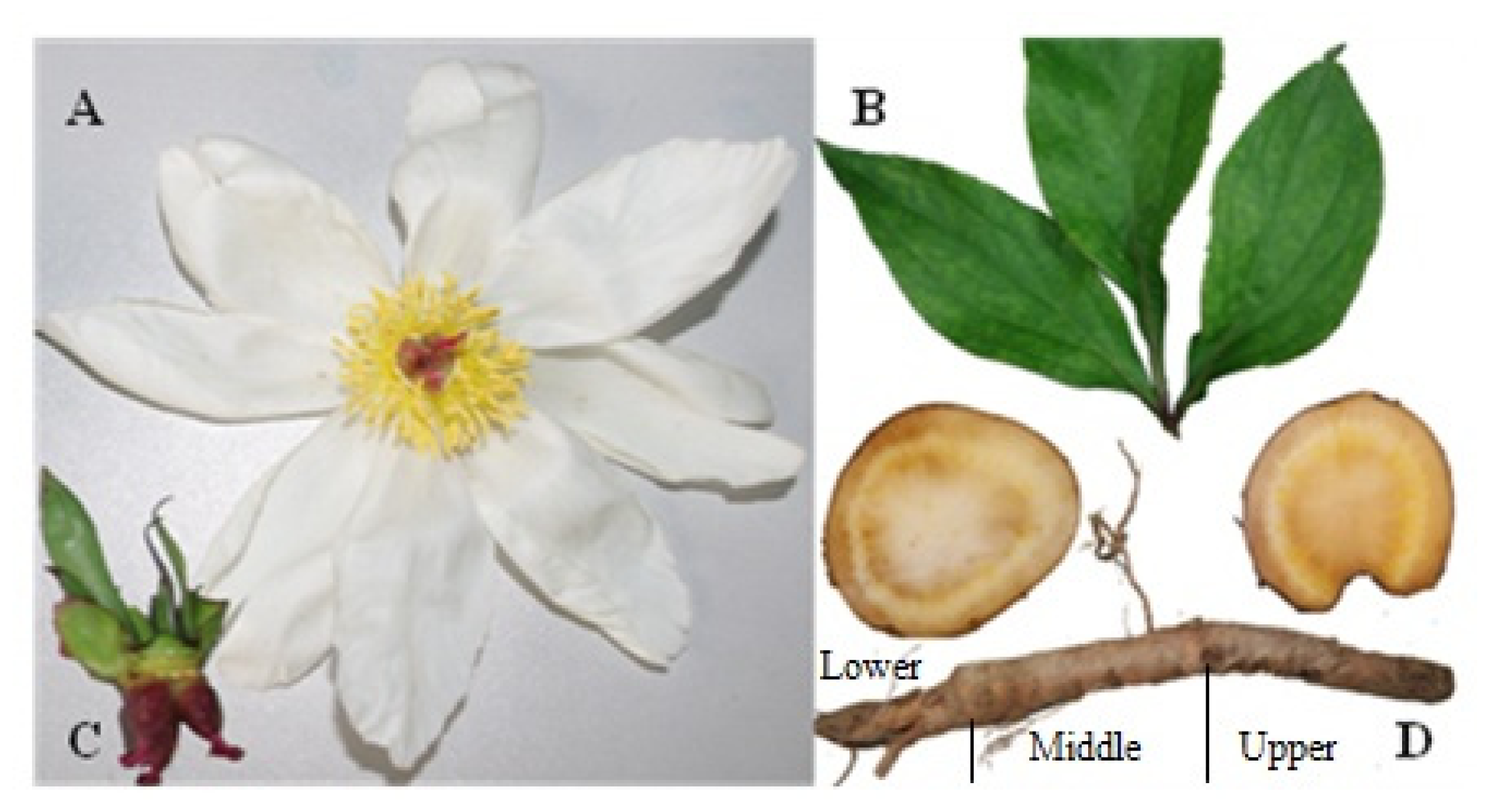
2.3. Tissue-Specific Accumulation of Active Compounds in P. lactiflora
2.4. Expression of Gallic Acid Biosynthesis-Related Genes in Different Tissues of P. lactiflora
2.5. Expression of Paeoniflorin Biosynthesis-Related Genes in Different Tissues of P. lactiflora
2.6. Correlation of Gene Expression and Active Compound Contents
3. Disscussion
3.1. Preferential Expression of Gene Families in Bark
3.2. Variations of Key Genes in Shikimate Biosynthesis and the MVA Pathway Lead to Active Compound Accumulation
3.3. Gene and Functional Diversity of Paeoniflorin and Gallic Acid Biosyntheses Pathway
4. Experimental Section
4.1. Plant Materials
4.2. Gene Prediction and KEGG Annotation
4.3. Sequence and Phylogenic Analyses
4.4. RNA Extraction and PCR Analysis
4.5. Chemical Analysis
4.6. Correlation Analysis
5. Conclusions
Supplementary Information
ijms-14-18502-s001.pdfAcknowledgments
Conflicts of Interest
References
- He, D.; Dai, S. Anti-inflammatory and immunomodulatory effects of Paeonia lactiflora Pall., a traditional Chinese herbal medicine. Front Pharmacol 2011, 2. [Google Scholar] [CrossRef]
- Peng, H.S.; Wang, D.Q. The development and evolution of differentiation between Radix Paeoniae Rubra and Radix Paeoniae Alba. Chin. J. Med. Hist 2007, 37, 133–136. [Google Scholar]
- Lin, H.C.; Ding, H.Y.; Wu, Y.C. Two novel compounds from Paeonia suffruticosa. J. Nat. Prod 1998, 61, 343–346. [Google Scholar]
- Zhang, X.; Wang, J.; Li, X. A study on the chemical constituents of Paeonia lactiflora Pall. J. Shenyang Pharm. Univ 2001, 18, 30–32. [Google Scholar]
- Tan, J.; Zhao, Q.; Yang, L.; Shang, Z.; Du, Z.; Yan, M. Chemical constituents in roots of Paeonia lactiflora. Chin. Tradit. Herb. Drugs 2010, 41, 1245–1248. [Google Scholar]
- Wang, R.; Lu, T.; Li, Y.W.; Hu, Y.J.; Wang, Z.T. A comparison on pharmacological actions between Radix Paeoniae Rubra and Radix Paeoniae Alba. China J. Exper. Trad. Med. Form 2010, 16, 113–114. [Google Scholar]
- Li, H.; Wang, S.; Zhang, B. Pharmacological action and pharmacokinetics of paeonol. Asia-Pacific Tradit. Med 2010, 6, 110–112. [Google Scholar]
- Lau, C.H.; Chan, C.M.; Chan, Y.W. Pharmacological investigations of the anti-diabetic effect of Cortex Moutan and its active component paeonol. Phytomedicine 2007, 14, 778–784. [Google Scholar]
- Sun, G.P.; Wan, X.; Xu, S.P.; Wang, H.; Liu, S.H.; Wang, Z.G. Antiproliferation and apoptosis induction of paeonol in human esophageal cancer cell lines. Dis. Esophaqus 2008, 21, 723–729. [Google Scholar]
- Huang, X.; Zhu, W. The involvement of mitochondrial phosphate transporter in accelerating bud dormancy release during chilling treatment of tree peony (Paeonia suffruticosa). Planta 2008, 228, 545–552. [Google Scholar]
- Zhao, D.; Tao, J.; Han, C.; Ge, J. Flower color diversity revealed by differential expression of flavonoid biosynthetic genes and flavonoid accumulation in herbaceous peony (Paeonia lactiflora Pall.). Mol. Biol. Rep 2012, 39, 11263–11275. [Google Scholar]
- Zhang, X.Y.; Li, X. Chemical research progress on white peony. Shenyang Pharm. Univ. Bulletin 2002, 19, 70–73. [Google Scholar]
- Kitagawa, I.; Yoshikawa, M.; Tsunaga, K. Moutanpi ingredients. Shyakugaku Zasshi 1979, 33, 171–177. [Google Scholar]
- Ren, Y.; Qu, W.; Liang, J. Progress on chemical constituents and anti-prostate cancer of Punica Granatum. Strait Pharm. J 2011, 23, 7–11. [Google Scholar]
- Djakpo, O.; Yao, W. Rhus Chinensis and Galla Chinensis—Folklore to modern evidence: Review. Phytother. Res 2010, 24, 1739–1747. [Google Scholar]
- Wei, H.; Zeng, F.J.; Lu, M.Y.; Tang, R.J. Studies on chemical constituents from the root of Coriaria nepalensis wall. Acta Pharm. Sin 1998, 33, 688–692. [Google Scholar]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol 2000, 132, 365–386. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994, 22, 4673–4680. [Google Scholar]
- Hittinger, C.T.; Carroll, S.B. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 2007, 449, 677–681. [Google Scholar]
- Ding, L.; Hofius, D.; Hajirezaei, M.R.; Fernie, A.R.; Bornke, F.; Sonnewald, U. Functional analysis of the essential bifunctional tobacco enzyme 3-dehydroquinate dehydratase/shikimate dehydrogenase in transgenic tobacco plants. J. Exp. Bot 2007, 8, 2053–2067. [Google Scholar]
- Lange, B.M.; Ketchum, R.E.; Croteau, R.B. Isoprenoid biosynthesis. Metabolite profiling of peppermint oil gland secretory cells and application to herbicide target analysis. Plant Physiol 2001, 127, 305–314. [Google Scholar]
- Ishiguro, S.; Nishimori, Y.; Yamada, M.; Saito, H.; Suzuki, T.; Nakagawa, T.; Miyake, H.; Okada, K.; Nakamura, K. The Arabidopsis FLAKY POLLEN1 gene encodes a 3-hydroxy-3-methylglutaryl-coenzyme A synthase required for development of tapetum-specific organelles and fertility of pollen grains. Plant Cell Physiol 2010, 51, 896–911. [Google Scholar]
- Yang, J.; Xian, M.; Su, S.; Zhao, G.; Nie, Q.; Jiang, X.; Zheng, Y.; Liu, W. Enhancing production of bio-isoprene using hybrid MVA pathway and isoprene synthase in E. coli. PLoS One 2012, 7, e33509. [Google Scholar]
- Herdendorf, T.J.; Miziorko, H.M. Phosphomevalonate kinase: Functional investigation of the recombinant human enzyme. Biochemistry 2006, 45, 3235–3242. [Google Scholar]
- Van Deenen, N.; Bachmann, A.L.; Schmidt, T.; Schaller, H.; Sand, J.; Prufer, D.; Schulze, G.C. Molecular cloning of mevalonate pathway genes from Taraxacum brevicorniculatum and functional characterisation of the key enzyme 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Mol. Biol. Rep 2012, 39, 4337–4349. [Google Scholar]
- Wang, H.; Nagegowda, D.A.; Rawat, R.; Bouvier-Nave, P.; Guo, D.; Bach, T.J.; Chye, M.L. Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant Biotechnol. J 2012, 10, 31–42. [Google Scholar]
- Ohno, S. Evolution by Gene Duplication; Springer-Verlag: New York, NY, USA, 1970; pp. 59–87. [Google Scholar]
- Rastogi, S.; Liberles, D. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol. Biol 2005, 5, 28. [Google Scholar]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000, 28, 27–30. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar]
- Burge, C.B.; Karlin, S. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol 1998, 8, 346–354. [Google Scholar]
- Bjellqvist, B.; Basse, B.; Olsen, E.; Celis, J.E. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 1994, 15, 529–539. [Google Scholar]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res 2005, 33, W116–W120. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 2011, 28, 2731–2739. [Google Scholar]
- Kumar, S.; Dudley, J.; Nei, M.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform 2008, 9, 299–306. [Google Scholar]

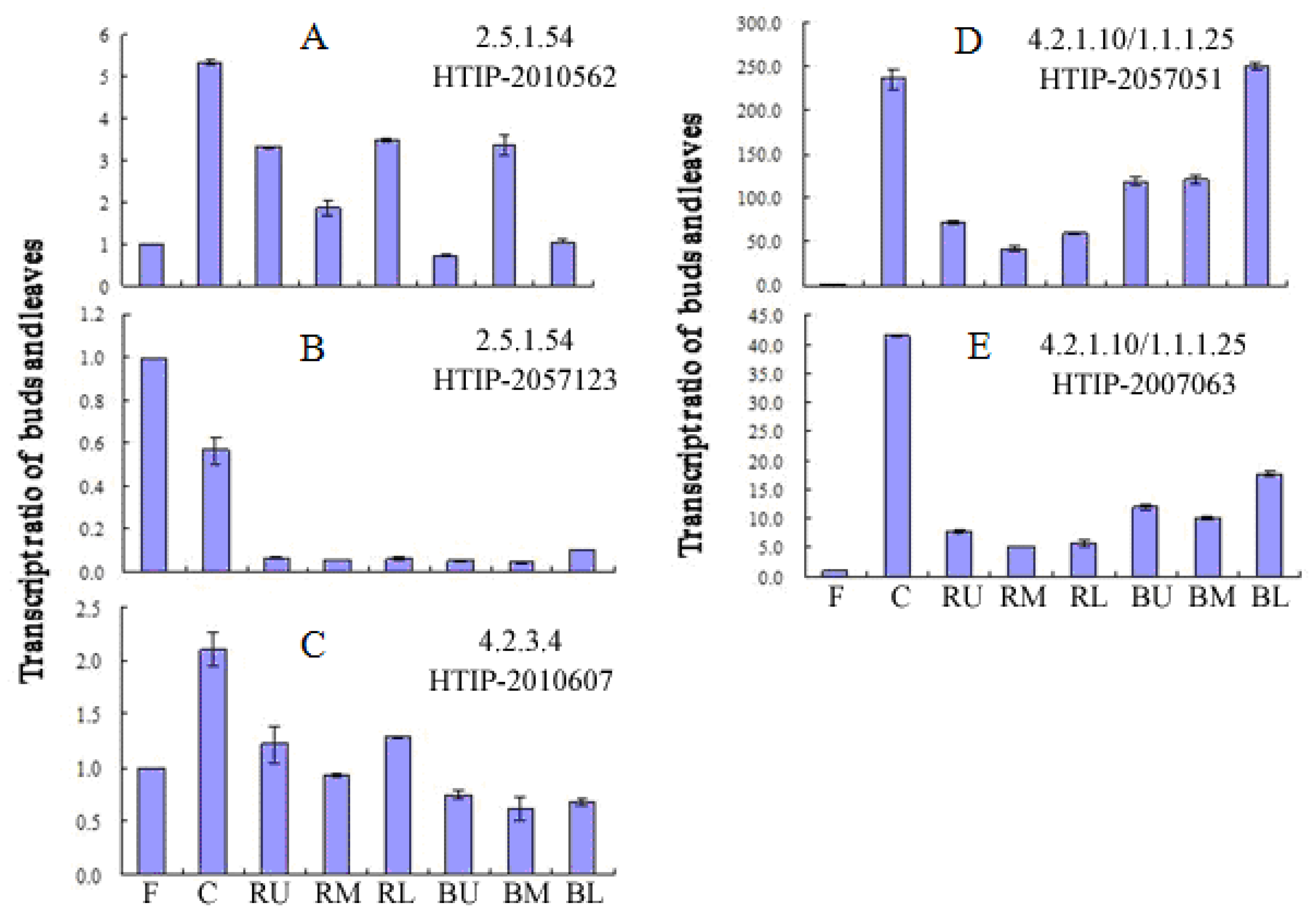
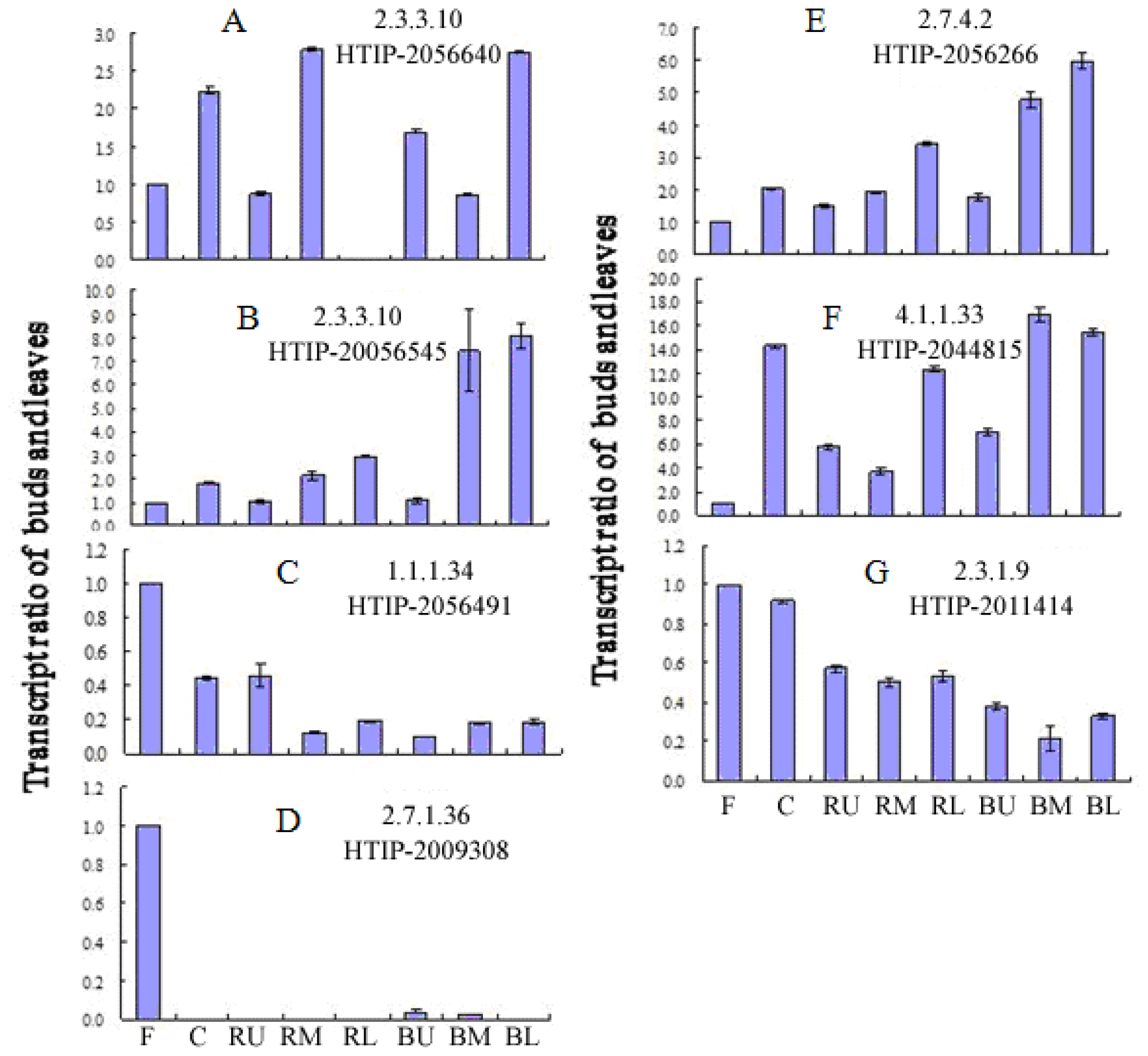
| Enzyme | Name | Contig No. | Length (bp) |
|---|---|---|---|
| 2.5.1.54 | PLDHS1 | HTIP-2010562 | 2105* |
| PLDHS2 | HTIP-2057123 | 2054 | |
| 4.2.3.4 | PLDHQS | HTIP-2010607 | 1790 |
| 4.2.1.10/ | PLaroDE1 | HTIP-2007063 | 1963 |
| 1.1.1.25 | PLaroDE2 | HTIP-2057051 | 1971 |
| 2.3.3.10 | PLHMGS1 | HTIP-2056545 | 1615* |
| PLHMGS2 | HTIP-2056640 | 1661* | |
| 1.1.1.34 | PLHMGCR | HTIP-2056491 | 1591 |
| 2.7.1.36 | PLMVK | HTIP-2009308 | 1464 |
| 2.7.4.2 | PLPMK | HTIP-2056266 | 1494 |
| 4.1.1.33 | PLMVD1 | HTIP-2044815 | 316 |
| PLMVD2 | HTIP-2049597 | 479 | |
| 2.3.1.9 | PLACAT | HTIP-2011414 | 1399* |
| PLDXPS1 | HTIP-2003301 | 2518 | |
| 2.2.1.7 | PLDXPS2 | HTIP-2003675 | 2488* |
| PLDXPS3 | HTIP-2057315 | 2338* | |
| 1.1.1.267 | PLDXR1 | HTIP-2000861 | 423 |
| PLDXR2 | HTIP-2006951 | 1040 | |
| 2.7.7.60 | PLIspD | HTIP-2008416 | 1345 |
| 2.7.1.148 | PLCMK | HTIP-2008205 | 1609* |
| 4.6.1.12 | PLIspF | HTIP-2054028 | 896* |
| 1.17.7.1 | PLHDS | HTIP-2007108 | 2665 |
| 1.17.7.2 | PLHDR | HTIP-2056592 | 1637 |
| 5.3.3.2 | PLIDI | HTIP-2055407 | 1185 |
| Name | Length (aa) | Molecular weight (KD) | Isoelectric point | Subcellular Localization | Signal peptide | Domains (# of domains) |
|---|---|---|---|---|---|---|
| DHS1 | 528 | 58.7 | 8.06 | Mit. | No | DAHP synthetase, class II(IPR002480) |
| HMGCS1 | 453 | 50.6 | 6.10 | Cytosol | No | Hydroxymethylglutaryl-coenzyme A synthase, active site(IPR000590) |
| HMGCS2 | 465 | 51.3 | 5.98 | Cytosol | No | Hydroxymethylglutaryl-coenzyme A synthase, active site(IPR000590) |
| ACAT | 405 | 41.4 | 6.15 | Cytosol | No | Thiolase (IPR002155) |
| DXPS2 | 714 | 77.0 | 6.60 | Chl. | No | Deoxyxylulose-5-phosphate synthase (IPR005477) |
| DXPS3 | 719 | 78.9 | 6.17 | Cytosol | No | Deoxyxylulose-5-phosphate synthase (IPR005477) |
| CMK | 403 | 44.5 | 6.13 | Chl. | No | 4-diphosphocytidyl-2C-methyl-d-erthritol kinase(IPR004424) |
| IspF | 231 | 25.0 | 7.89 | Chl. | No | 2-C-methyl-d-erythritol 2,4-cyclodophosphate synthase (IPR003526) |
| Enzyme | Gene | Peony lactone glycosides | Paeoniflorin | Benzoic acid | Benzoyl paeoniflorin | Paeonol |
|---|---|---|---|---|---|---|
| Shikimate biosynthesis | ||||||
| 2.5.1.54 | 2010562 | −0.40 | −0.71 | −0.36 | −0.38 | −0.60 |
| 2057123 | −0.11 | 0.70 | 0.68 | 0.76 | 0.48 | |
| 4.2.3.4 | 2010607 | 0.04 | −0.51 | −0.64 | −0.57 | −0.40 |
| 4.2.1.10/ | 2057051 | −0.05 | 0.72 | 0.95 | 0.94 | 0.68 |
| 1.1.1.25 | 2007063 | 0.14 | 0.71 | 0.88 | 0.86 | 0.80 |
| MVA pathway | ||||||
| 2.3.3.10 | 2056545 | −0.63 | 0.38 | 0.88 | 0.83 | 0.07 |
| 2056640 | −0.02 | 0.89 | 0.45 | 0.50 | 0.30 | |
| 1.1.1.34 | 2056491 | 0.15 | −0.08 | −0.12 | −0.08 | 0.15 |
| 2.7.1.36 | 2009308 | 0.55 | −0.20 | −0.10 | −0.19 | 0.38 |
| 2.7.4.2 | 2056266 | −0.62 | 0.36 | 0.88 | 0.84 | 0.10 |
| 2.3.1.9 | 2011414 | 0.17 | −0.24 | −0.64 | −0.55 | −0.28 |
| 4.1.1.33 | 2044815 | −0.50 | 0.01 | 0.70 | 0.62 | 0.06 |
| MEP/DXP pathway | ||||||
| 2.2.1.7 | 2003675 | 0.12 | −0.22 | −0.55 | −0.47 | −0.23 |
| 2003301 | −0.19 | −0.14 | 0.07 | 0.12 | −0.06 | |
| 2057315 | 0.19 | −0.72 | −0.75 | −0.76 | −0.44 | |
| 1.1.1.267 | 2000861 | −0.22 | −0.64 | −0.38 | −0.37 | −0.49 |
| 2006951 | −0.21 | −0.57 | −0.51 | −0.47 | −0.54 | |
| 2.7.7.60 | 2008416 | 0.71 | −0.37 | −0.43 | −0.48 | 0.27 |
| 2.7.1.148 | 2008205 | −0.07 | −0.49 | −0.38 | −0.33 | −0.29 |
| 4.6.1.12 | 2054028 | −0.05 | −0.48 | −0.42 | −0.37 | −0.31 |
| 1.17.7.1 | 2007108 | −0.03 | −0.55 | −0.43 | −0.39 | −0.32 |
| 1.17.7.2 | 2056592 | −0.15 | −0.48 | −0.45 | −0.39 | −0.42 |
| 5.3.3.2 | 2055407 | −0.57 | −0.30 | 0.09 | 0.11 | −0.45 |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yuan, Y.; Yu, J.; Jiang, C.; Li, M.; Lin, S.; Wang, X.; Huang, L. Functional Diversity of Genes for the Biosynthesis of Paeoniflorin and Its Derivatives in Paeonia. Int. J. Mol. Sci. 2013, 14, 18502-18519. https://doi.org/10.3390/ijms140918502
Yuan Y, Yu J, Jiang C, Li M, Lin S, Wang X, Huang L. Functional Diversity of Genes for the Biosynthesis of Paeoniflorin and Its Derivatives in Paeonia. International Journal of Molecular Sciences. 2013; 14(9):18502-18519. https://doi.org/10.3390/ijms140918502
Chicago/Turabian StyleYuan, Yuan, Jun Yu, Chao Jiang, Minhui Li, Shufang Lin, Xumin Wang, and Luqi Huang. 2013. "Functional Diversity of Genes for the Biosynthesis of Paeoniflorin and Its Derivatives in Paeonia" International Journal of Molecular Sciences 14, no. 9: 18502-18519. https://doi.org/10.3390/ijms140918502




