High-Level Overproduction of Thermobifida Enzyme in Streptomyces lividans Using a Novel Expression Vector
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characteristics of pZRJ362
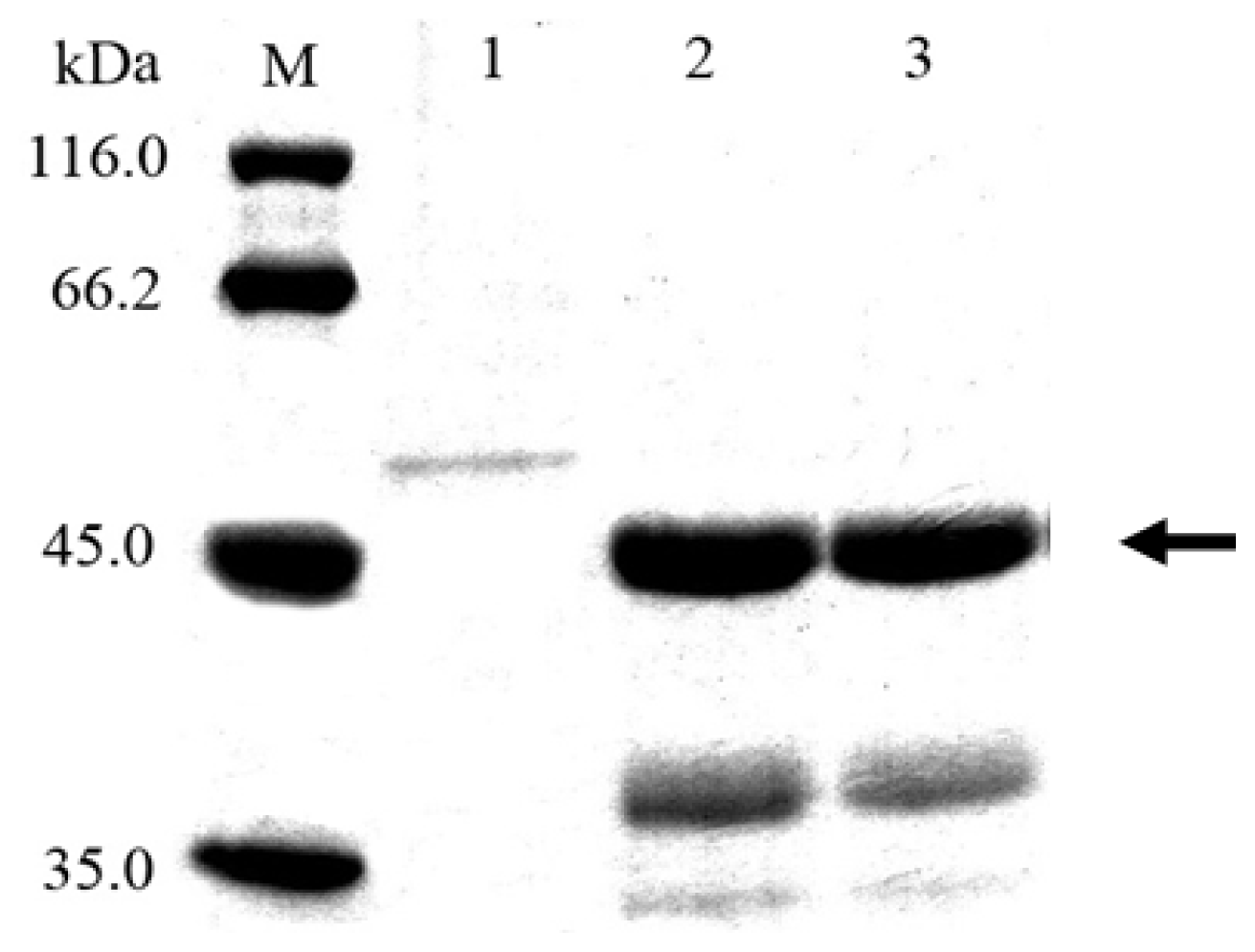
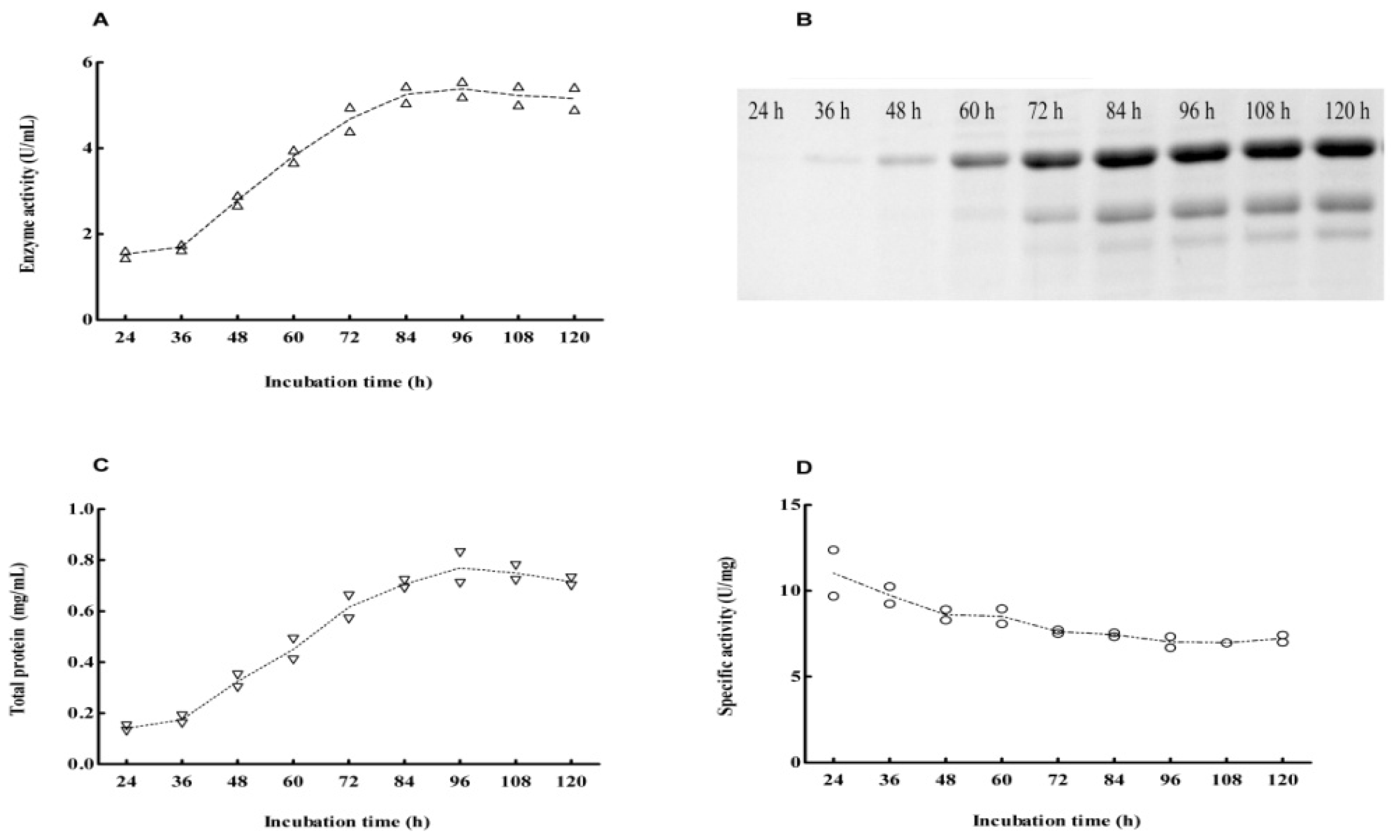
2.2. Cloning and Expression of Cel6A in Streptomyces lividans
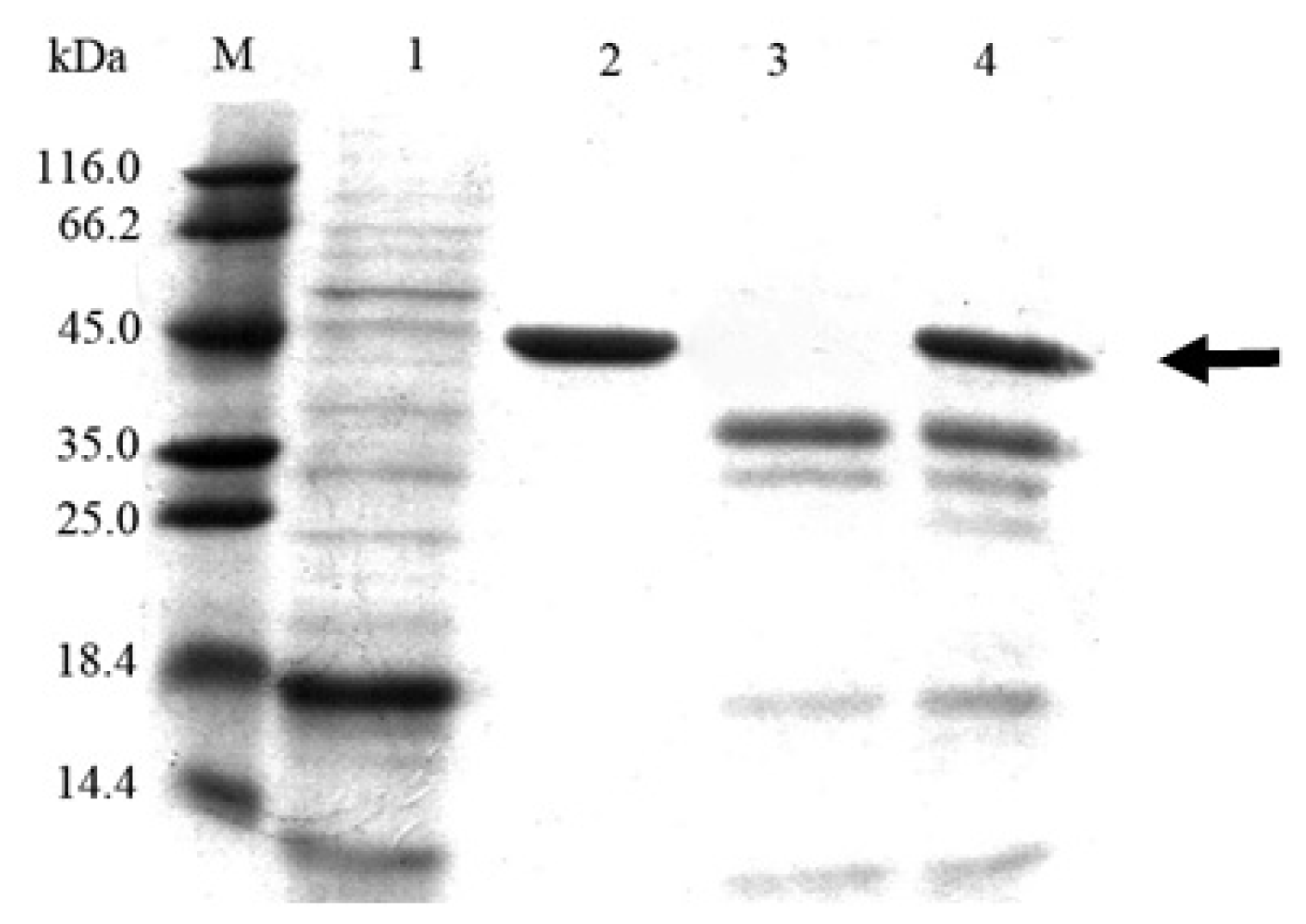
2.3. Cloning and Expression of Cel6A in Pichia pastoris
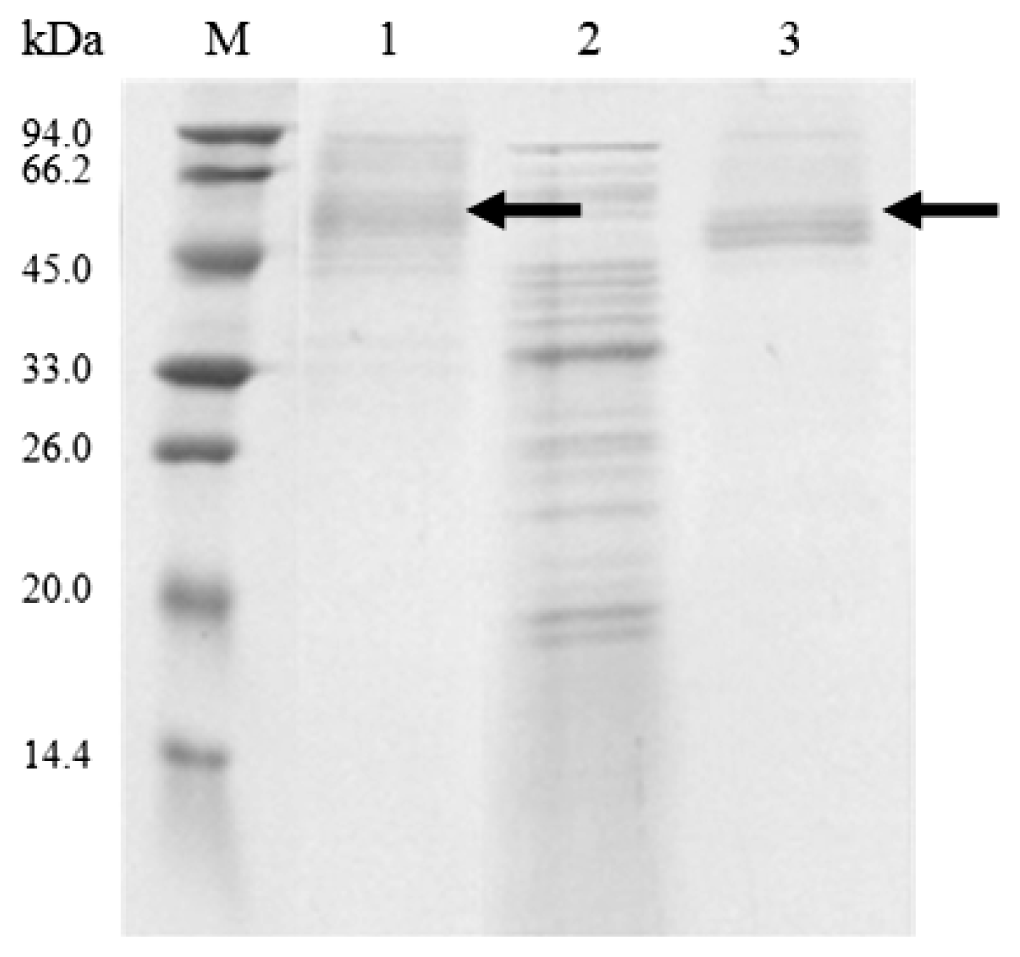
2.4. Expression Pattern of Cel6A in Streptomyces lividans versus Pichia pastoris
3. Experimental Section
3.1. Strains, Plasmids and Culture Conditions
3.2. Construction of Expression Plasmid pZRJ362
3.3. Cloning of Endoglucanase Gene cel6A in Streptomyces lividans
3.4. Cloning of Endoglucanase Gene cel6A in Pichia pastoris
3.5. Production and Purification of Cel6A
3.6. Endoglucanase Activity Assays
3.7. SDS-PAGE and Endoglucanase Activity Staining
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Ogino, C.; Kanemasu, M.; Hayashi, Y.; Kondo, A.; Shimizu, N.; Tokuyama, S.; Tahara, Y.; Kuroda, S.; Tanizawa, K.; Fukuda, H. Over-expression system for secretory phospholipase D by Streptomyces lividans. Appl. Microbiol. Biot 2004, 64, 823–828. [Google Scholar]
- Henne, A.; Brüggemann, H.; Raasch, C.; Wiezer, A.; Hartsch, T.; Liesegang, H.; Johann, A.; Lienard, T.; Gohl, O.; Martinez-Arias, R. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol 2004, 22, 547–553. [Google Scholar]
- Wilson, D.B. Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem. Rec 2004, 4, 72–82. [Google Scholar]
- Xie, F.; Chao, Y.; Yang, X.; Yang, J.; Xue, Z.; Luo, Y.; Qian, S. Purification and characterization of four keratinases produced by Streptomyces sp. Strain 16 in native human foot skin medium. Bioresource Technol 2010, 101, 344–350. [Google Scholar]
- Mehta, V.; Thumar, J.; Singh, S. Production of alkaline protease from an alkaliphilic actinomycete. Bioresource Technol 2006, 97, 1650–1654. [Google Scholar]
- Thumar, J.T.; Singh, S.P. Secretion of an alkaline protease from a salt-tolerant and alkaliphilic, Streptomyces clavuligerus strain Mit-1. Braz. J. Microbiol 2007, 38, 766–772. [Google Scholar]
- Yang, C.-H.; Huang, Y.-C.; Chen, C.-Y.; Wen, C.-Y. Expression of Thermobifida fusca thermostable raw starch digesting alpha-amylase in Pichia pastoris and its application in raw sago starch hydrolysis. J. Ind. Microbiol. Biot 2010, 37, 401–406. [Google Scholar]
- Yang, C.-H.; Liu, W.-H. Cloning and characterization of a maltotriose-producing α-amylase gene from Thermobifida fusca. J. Ind. Microbiol. Biot 2007, 34, 325–330. [Google Scholar]
- Yang, C.-H.; Yang, S.-F.; Liu, W.-H. Production of xylooligosaccharides from xylans by extracellular xylanases from Thermobifida fusca. J. Agric. Food Chem 2007, 55, 3955–3959. [Google Scholar]
- Zhou, J.; Gao, Y.; Dong, Y.; Tang, X.; Li, J.; Xu, B.; Mu, Y.; Wu, Q.; Huang, Z. A novel xylanase with tolerance to ethanol, salt, protease, sds, heat, and alkali from actinomycete Lechevalieria sp. HJ3. J. Ind. Microbiol. Biot 2012, 39, 965–975. [Google Scholar]
- Kumar, A.; Gupta, R.; Shrivastava, B.; Khasa, Y.P.; Kuhad, R.C. Xylanase production from an alkalophilic actinomycete isolate Streptomyces sp. RCK-2010, its characterization and application in saccharification of second generation biomass. J. Mol. Catal. B 2012, 74, 170–177. [Google Scholar]
- Sriyapai, T.; Somyoonsap, P.; Matsui, K.; Kawai, F.; Chansiri, K. Cloning of a thermostable xylanase from Actinomadura sp. S14 and its expression in Escherichia coli and Pichia pastoris. J. Biosci. Bioeng 2011, 111, 528–536. [Google Scholar]
- Díaz, M.; Ferreras, E.; Moreno, R.; Yepes, A.; Berenguer, J.; Santamaría, R. High-level overproduction of Thermus enzymes in Streptomyces lividans. Appl. Microbiol. Biot 2008, 79, 1001–1008. [Google Scholar]
- Ghangas, G.S.; Wilson, D.B. Cloning of the Thermomonospora fusca endoglucanase E2 gene in Streptomyces lividans: Affinity purification and functional domains of the cloned gene product. Appl. Environ. Microb 1988, 54, 2521–2526. [Google Scholar]
- Iwasaki, Y.; Nakano, H.; Yamane, T. Phospholipase D from Streptomyces antibioticus: Cloning, sequencing, expression, and relationship to other phospholipases. Appl. Microbiol. Biot 1994, 42, 290–299. [Google Scholar]
- Wilson, D.B. Biochemistry and genetics of actinomycete cellulases. Crit. Rev. Biotechnol 1992, 12, 45–63. [Google Scholar]
- Yang, C.-H.; Lin, K.-I.; Chen, G.-H.; Chen, Y.-F.; Chen, C.-Y.; Chen, W.-L.; Huang, Y.-C. Constitutive expression of Thermobifida fusca thermostable acetylxylan esterase gene in Pichia pastoris. Int. J. Mol. Sci 2010, 11, 5143–5151. [Google Scholar]
- Sianidis, G.; Pozidis, C.; Becker, F.; Vrancken, K.; Sjoeholm, C.; Karamanou, S.; Takamiya-Wik, M.; Van Mellaert, L.; Schaefer, T.; Anné, J. Functional large-scale production of a novel Jonesia sp. Xyloglucanase by heterologous secretion from Streptomyces lividans. J. Biotechnol 2006, 121, 498–507. [Google Scholar]
- Cheng, Y.-F.; Yang, C.-H.; Liu, W.-H. Cloning and expression of Thermobifida xylanase gene in the methylotrophic yeast Pichia pastoris. Enzyme Microb. Tech 2005, 37, 541–546. [Google Scholar]
- Whittaker, M.M.; Whittaker, J.W. Streptomyces coelicolor oxidase (sco2837p): A new free radical metalloenzyme secreted by Streptomyces coelicolor A3 (2). Arch. Biochem. Biophys 2006, 452, 108–118. [Google Scholar]
- Kim, T.; Lei, X.G. Expression and characterization of a thermostable serine protease (tfpa) from Thermomonospora fusca YX in Pichia pastoris. Appl. Microbiol. Biot 2005, 68, 355–359. [Google Scholar]
- Nakashima, N.; Mitani, Y.; Tamura, T. Actinomycetes as host cells for production of recombinant proteins. Microb. Cell Fact 2005, 4, 7. [Google Scholar]
- Bachmann, S.L.; McCarthy, A.J. Purification and cooperative activity of enzymes constituting the xylan-degrading system of Thermomonospora fusca. Appl. Environ. Microb 1991, 57, 2121–2130. [Google Scholar]
- Lykidis, A.; Mavromatis, K.; Ivanova, N.; Anderson, I.; Land, M.; DiBartolo, G.; Martinez, M.; Lapidus, A.; Lucas, S.; Copeland, A. Genome sequence and analysis of the soil cellulolytic actinomycete Thermobifida fusca YX. J. Bacteriol 2007, 189, 2477–2486. [Google Scholar]
- Noda, S.; Ito, Y.; Shimizu, N.; Tanaka, T.; Ogino, C.; Kondo, A. Over-production of various secretory-form proteins in Streptomyces lividans. Protein Expres. Purif 2010, 73, 198–202. [Google Scholar]
- Miller, G.L.; Blum, R.; Glennon, W.E.; Burton, A.L. Measurement of carboxymethylcellulase activity. Anal. Biochem 1960, 1, 127–132. [Google Scholar]
- Sun, X.; Liu, Z.; Zheng, K.; Song, X.; Qu, Y. The composition of basal and induced cellulase systems in Penicillium decumbens under induction or repression conditions. Enzyme Microb. Tech 2008, 42, 560–567. [Google Scholar]


© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, J.-X.; Zhao, L.-M.; Wu, R.-J.; Zheng, Z.-J.; Zhang, R.-J. High-Level Overproduction of Thermobifida Enzyme in Streptomyces lividans Using a Novel Expression Vector. Int. J. Mol. Sci. 2013, 14, 18629-18639. https://doi.org/10.3390/ijms140918629
Li J-X, Zhao L-M, Wu R-J, Zheng Z-J, Zhang R-J. High-Level Overproduction of Thermobifida Enzyme in Streptomyces lividans Using a Novel Expression Vector. International Journal of Molecular Sciences. 2013; 14(9):18629-18639. https://doi.org/10.3390/ijms140918629
Chicago/Turabian StyleLi, Jun-Xia, Long-Mei Zhao, Ru-Juan Wu, Zhao-Jun Zheng, and Ri-Jun Zhang. 2013. "High-Level Overproduction of Thermobifida Enzyme in Streptomyces lividans Using a Novel Expression Vector" International Journal of Molecular Sciences 14, no. 9: 18629-18639. https://doi.org/10.3390/ijms140918629



