Small Molecules Present in the Cerebrospinal Fluid Metabolome Influence Superoxide Dismutase 1 Aggregation
Abstract
:1. Introduction
2. Results and Discussion
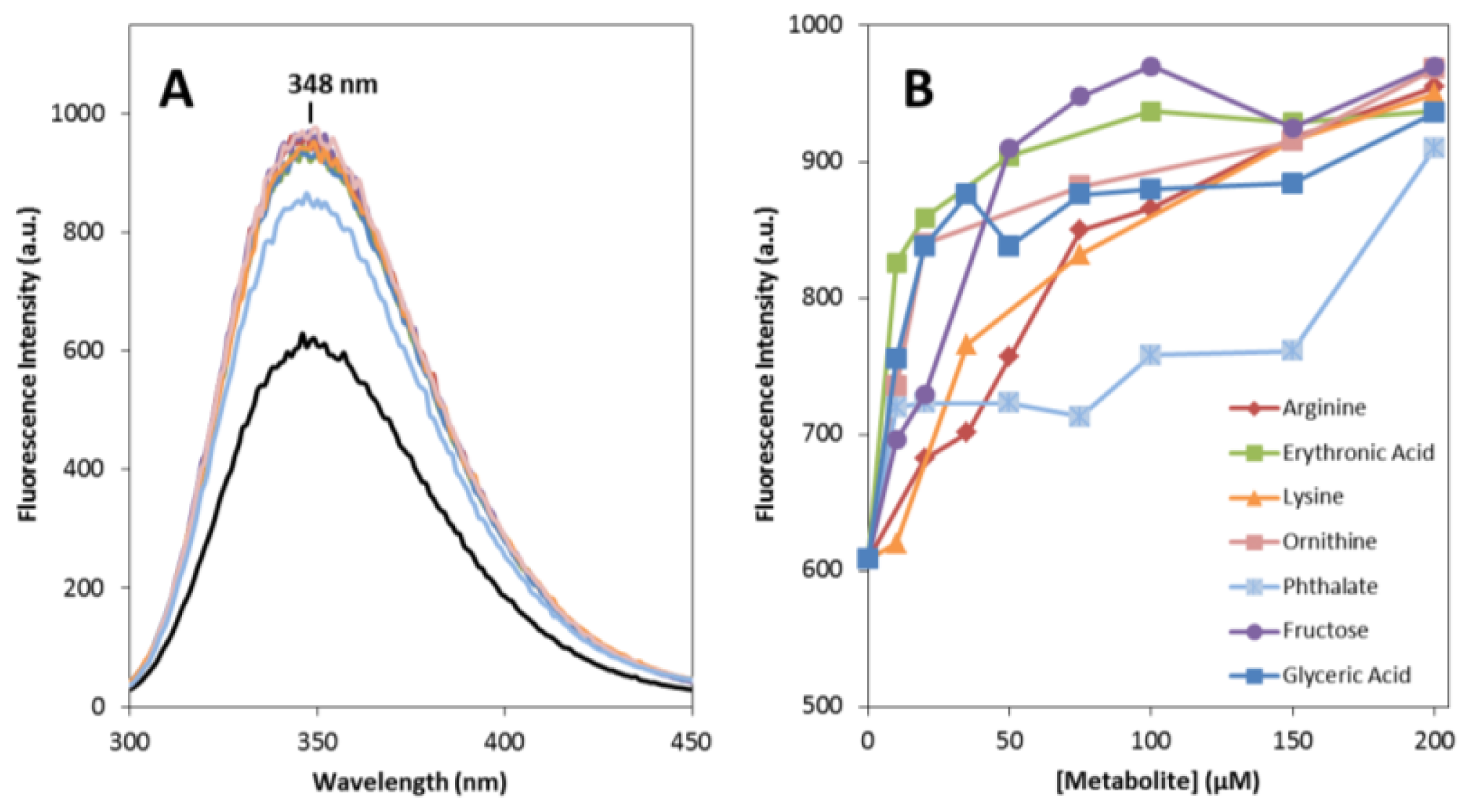
2.1. Effects on apoSOD1 Structure and Dynamics
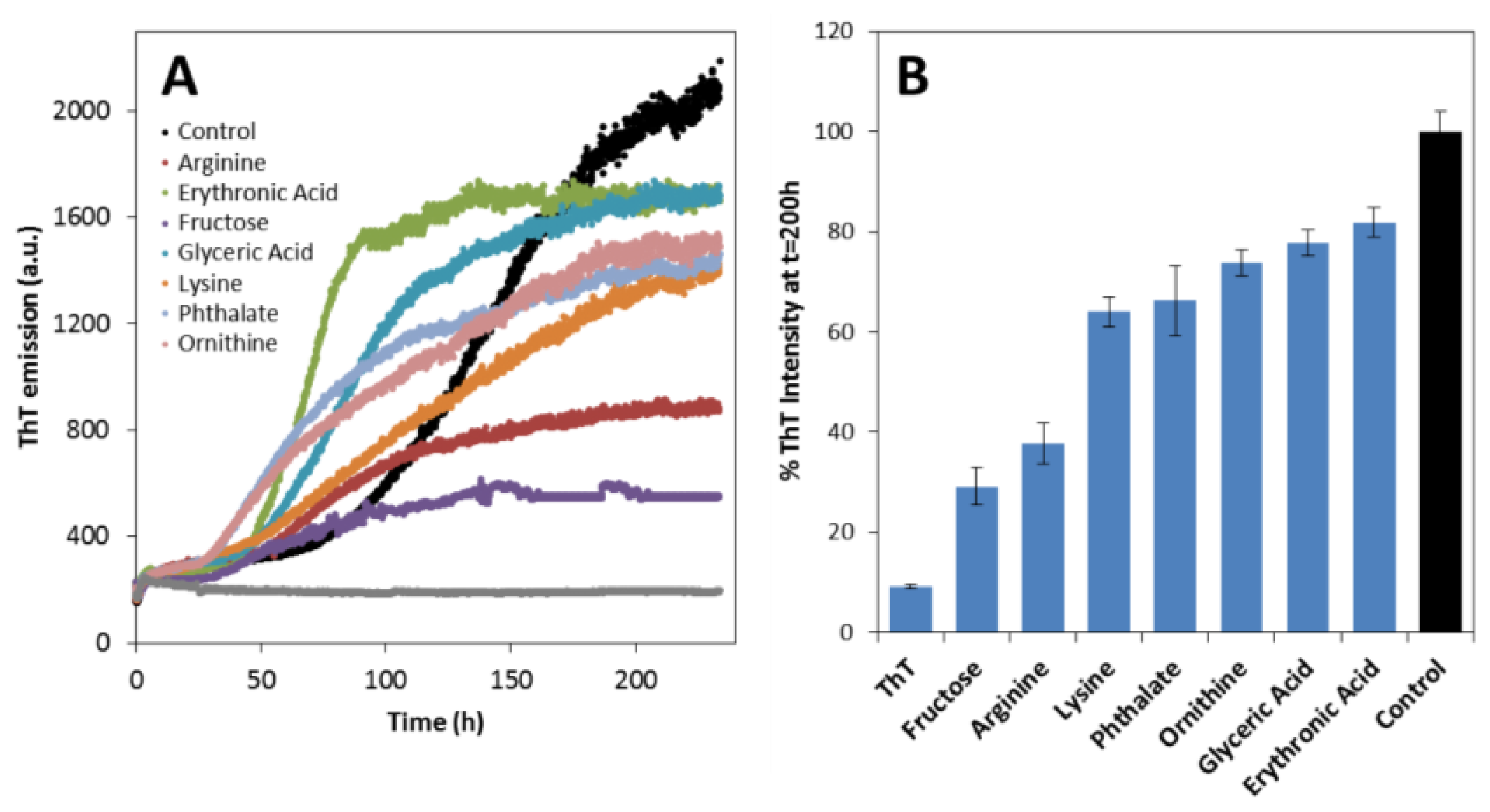
2.2. Effects on apoSOD1 Aggregation Kinetics
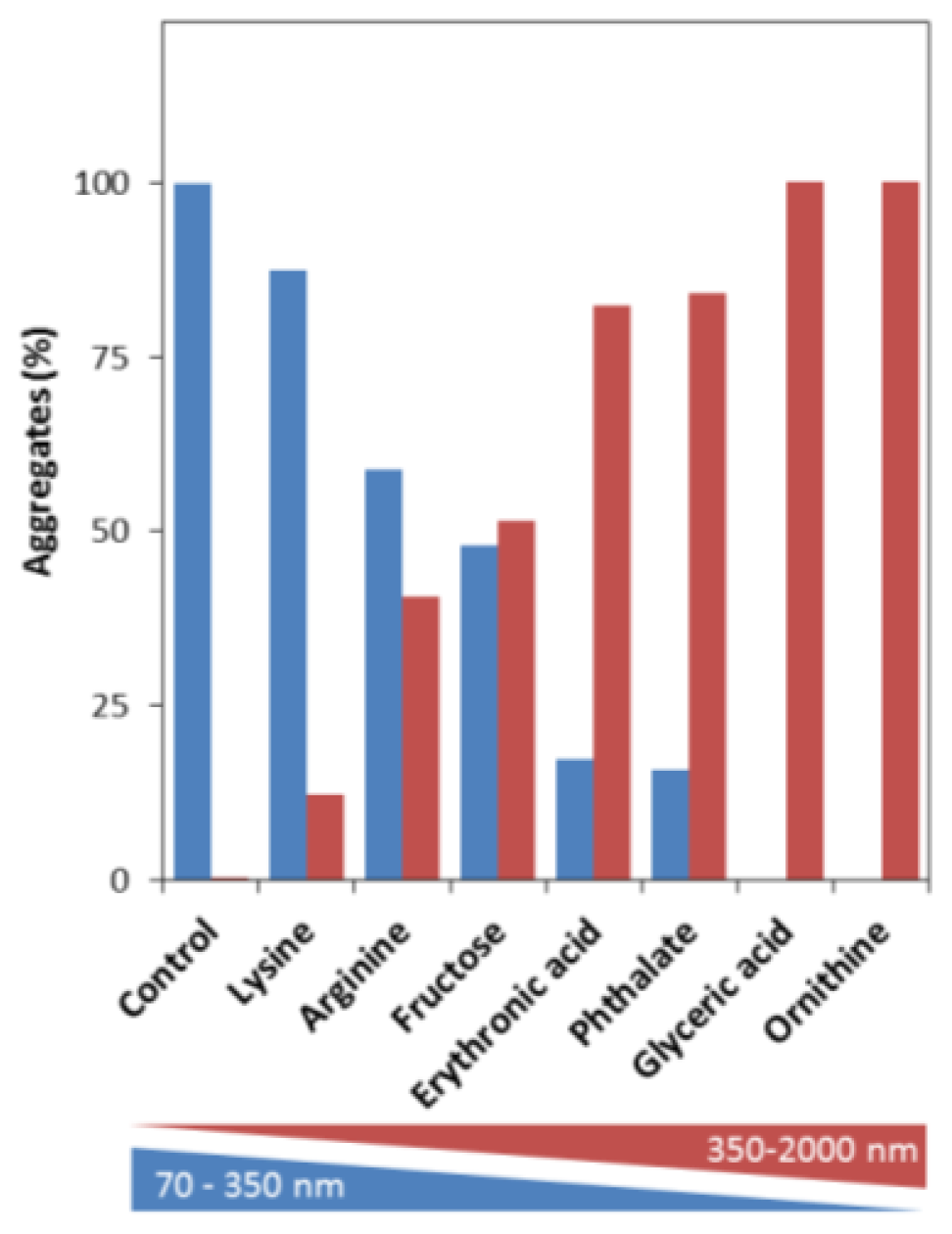
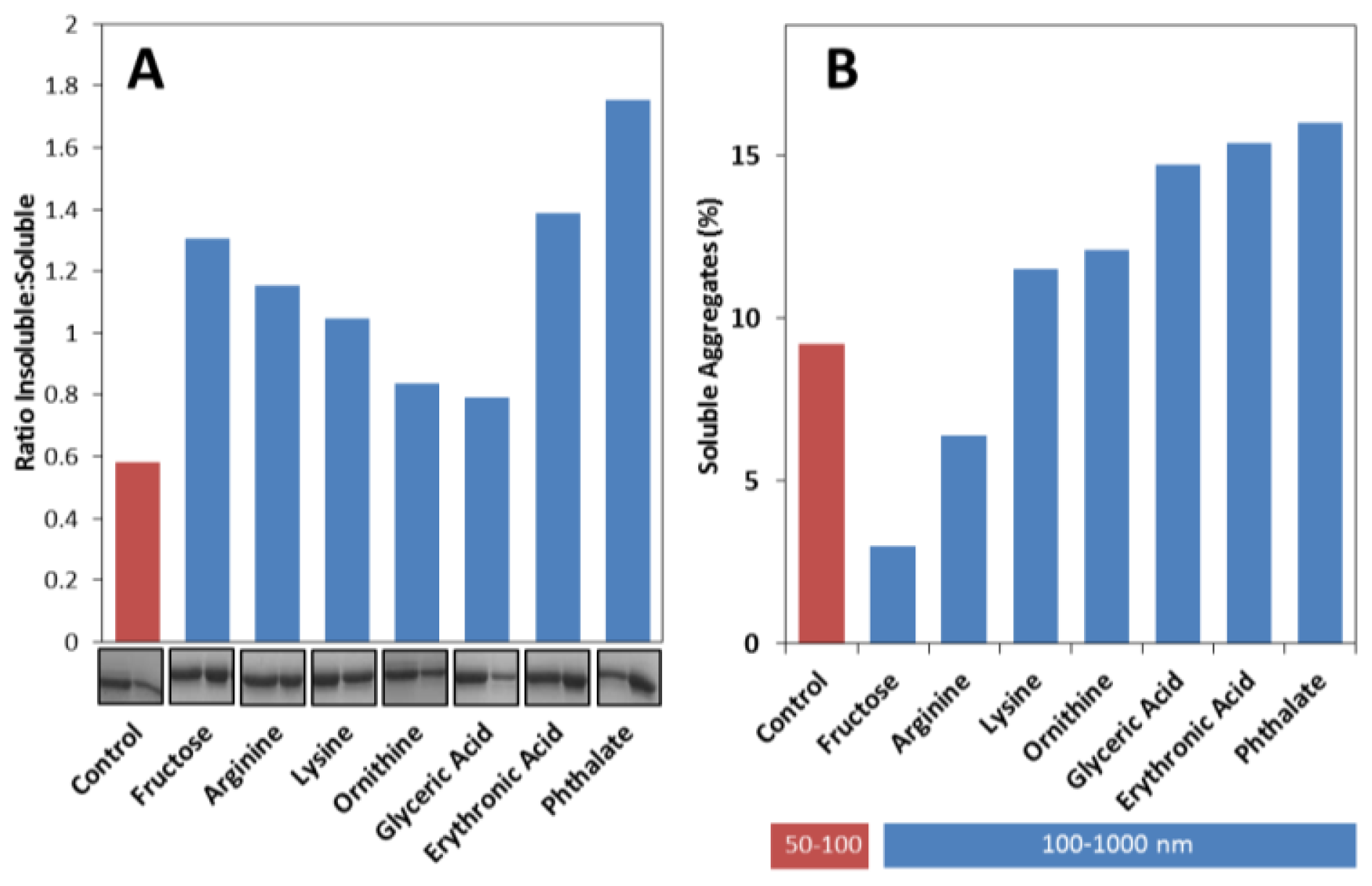
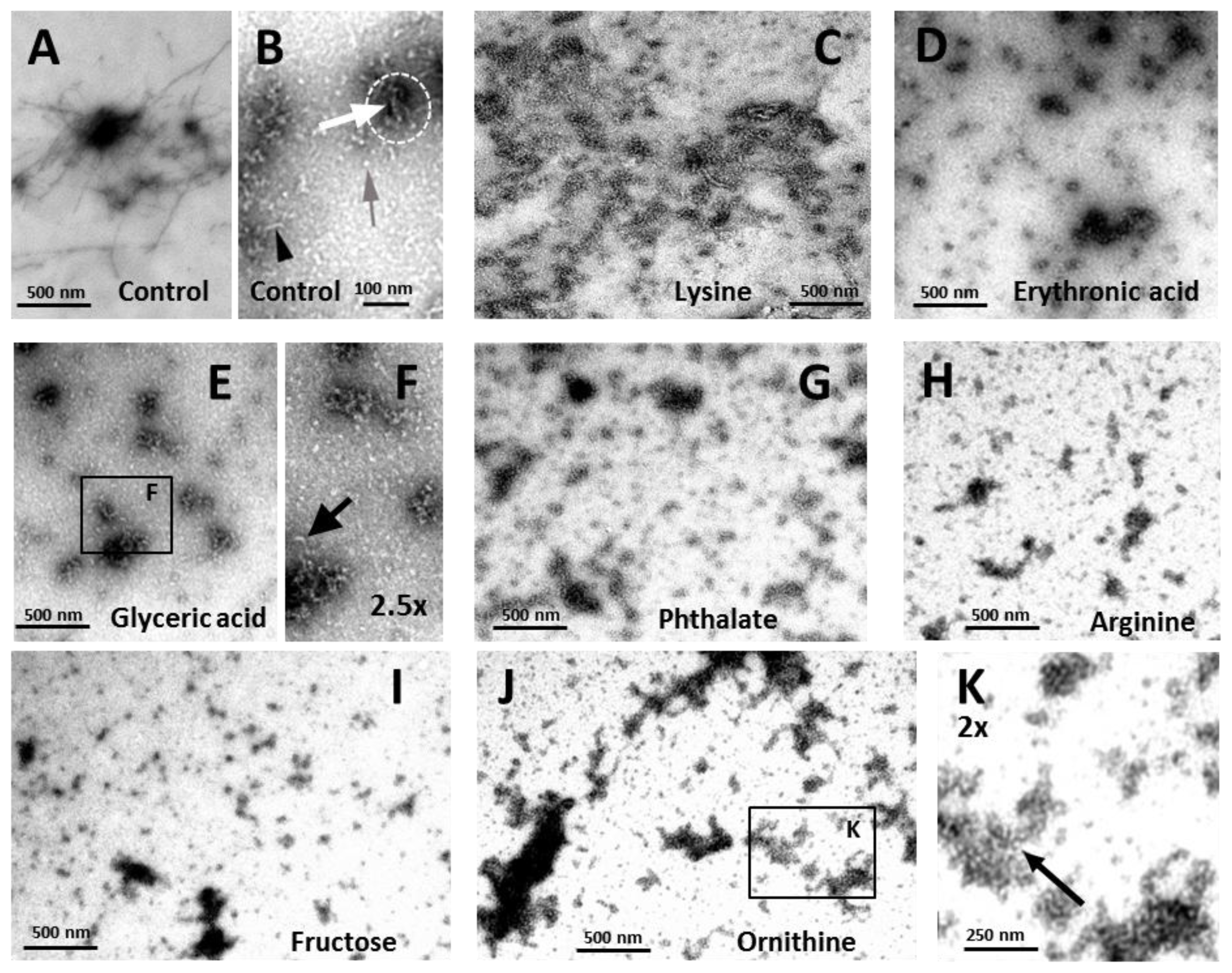
2.3. Effects on the Size, Solubility, and Morphology of apoSOD1 Aggregates
3. Experimental Section
3.1. Chemicals and Proteins
3.2. Fluorescence Spectroscopy
3.3. Circular Dichroism (CD)
3.4. Differential Scanning Fluorimetry
3.5. Trypsin Limited Proteolysis
3.6. SOD1 Aggregation Assays
3.7. Dynamic Light Scattering (DLS)
3.8. Nanoparticle Tracking Analysis (NTA)
3.9. Quantification of Insoluble Aggregates
3.10. Transmission Electron Microscopy
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Kato, S.; Takikawa, M.; Nakashima, K.; Hirano, A.; Cleveland, D.W.; Kusaka, H.; Shibata, N.; Kato, M.; Nakano, I.; Ohama, E. New consensus research on neuropathological aspects of familial amyotrophic lateral sclerosis with superoxide dismutase 1 (sod1) gene mutations: Inclusions containing sod1 in neurons and astrocytes. Amyotrophic Lateral Scler. Other Mot. Neuron Disord 2000, 1, 163–184. [Google Scholar]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.X.; et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar]
- Forsberg, K.; Jonsson, P.A.; Andersen, P.M.; Bergemalm, D.; Graffmo, K.S.; Hultdin, M.; Jacobsson, J.; Rosquist, R.; Marklund, S.L.; Brannstrom, T. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic als patients. PLoS One 2010, 5, e11552. [Google Scholar]
- Shibata, N.; Asayama, K.; Hirano, A.; Kobayashi, M. Immunohistochemical study on superoxide dismutases in spinal cords from autopsied patients with amyotrophic lateral sclerosis. Dev. Neurosci 1996, 18, 492–498. [Google Scholar]
- Forsberg, K.; Andersen, P.M.; Marklund, S.L.; Brannstrom, T. Glial nuclear aggregates of superoxide dismutase-1 are regularly present in patients with amyotrophic lateral sclerosis. Acta Neuropathol 2011, 121, 623–634. [Google Scholar]
- Kabashi, E.; Valdmanis, P.N.; Dion, P.; Rouleau, G.A. Oxidized/misfolded superoxide dismutase-1: The cause of all amyotrophic lateral sclerosis? Ann. Neurol 2007, 62, 553–559. [Google Scholar]
- Gruzman, A.; Wood, W.L.; Alpert, E.; Prasad, M.D.; Miller, R.G.; Rothstein, J.D.; Bowser, R.; Hamilton, R.; Wood, T.D.; Cleveland, D.W.; et al. Common molecular signature in sod1 for both sporadic and familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2007, 104, 12524–12529. [Google Scholar]
- Watanabe, M.; Dykes-Hoberg, M.; Culotta, V.C.; Price, D.L.; Wong, P.C.; Rothstein, J.D. Histological evidence of protein aggregation in mutant sod1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol. Dis 2001, 8, 933–941. [Google Scholar]
- Bruijn, L.I.; Houseweart, M.K.; Kato, S.; Anderson, K.L.; Anderson, S.D.; Ohama, E.; Reaume, A.G.; Scott, R.W.; Cleveland, D.W. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 1998, 281, 1851–1854. [Google Scholar]
- Ilieva, H.; Polymenidou, M.; Cleveland, D.W. Non-cell autonomous toxicity in neurodegenerative disorders: Als and beyond. J. Cell Biol 2009, 187, 761–772. [Google Scholar]
- Felbecker, A.; Camu, W.; Valdmanis, P.N.; Sperfeld, A.D.; Waibel, S.; Steinbach, P.; Rouleau, G.A.; Ludolph, A.C.; Andersen, P.M. Four familial als pedigrees discordant for two SOD1 mutations: Are all SOD1 mutations pathogenic? J. Neurol. Neurosurg. Psychiatry 2010, 81, 572–577. [Google Scholar]
- Roberts, K.; Zeineddine, R.; Corcoran, L.; Li, W.; Campbell, I.L.; Yerbury, J.J. Extracellular aggregated Cu/Zn superoxide dismutase activates microglia to give a cytotoxic phenotype. Glia 2013, 61, 409–419. [Google Scholar]
- Munch, C.; O’Brien, J.; Bertolotti, A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc. Natl. Acad. Sci. USA 2011, 108, 3548–3553. [Google Scholar]
- Wuolikainen, A.; Andersen, P.M.; Moritz, T.; Marklund, S.L.; Antti, H. Als patients with mutations in the sod1 gene have an unique metabolomic profile in the cerebrospinal fluid compared with als patients without mutations. Mol. Genet. Metab 2012, 105, 472–478. [Google Scholar]
- Tarasiuk, J.; Kulakowska, A.; Drozdowski, W.; Kornhuber, J.; Lewczuk, P. Csf markers in amyotrophic lateral sclerosis. J. Neural Transm 2012, 119, 747–757. [Google Scholar]
- Wuolikainen, A.; Moritz, T.; Marklund, S.L.; Antti, H.; Andersen, P.M. Disease-related changes in the cerebrospinal fluid metabolome in amyotrophic lateral sclerosis detected by gc/tofms. PLoS One 2011, 6, e17947. [Google Scholar]
- Niessen, H.G.; Debska-Vielhaber, G.; Sander, K.; Angenstein, F.; Ludolph, A.C.; Hilfert, L.; Willker, W.; Leibfritz, D.; Heinze, H.J.; Kunz, W.S.; et al. Metabolic progression markers of neurodegeneration in the transgenic g93a-sod1 mouse model of amyotrophic lateral sclerosis. Eur. J. Neurosci 2007, 25, 1669–1677. [Google Scholar]
- Mondola, P.; Ruggiero, G.; Seru, R.; Damiano, S.; Grimaldi, S.; Garbi, C.; Monda, M.; Greco, D.; Santillo, M. The Cu,Zn superoxide dismutase in neuroblastoma SK-N-BE cells is exported by a microvesicles dependent pathway. Brain Res. Mol. Brain Res 2003, 110, 45–51. [Google Scholar]
- Urushitani, M.; Sik, A.; Sakurai, T.; Nukina, N.; Takahashi, R.; Julien, J.P. Chromogranin-mediated secretion of mutant superoxide dismutase proteins linked to amyotrophic lateral sclerosis. Nat. Neurosci 2006, 9, 108–118. [Google Scholar]
- Jacobsson, J.; Jonsson, P.A.; Andersen, P.M.; Forsgren, L.; Marklund, S.L. Superoxide dismutase in csf from amyotrophic lateral sclerosis patients with and without Cu/Zn-superoxide dismutase mutations. Brain 2001, 124, 1461–1466. [Google Scholar]
- Zetterström, P.; Andersen, P.M.; Brännström, T.; Marklund, S.L. Misfolded superoxide dismutase-1 in CSF from amyotrophic lateral sclerosis patients. J. Neurochem 2011, 117, 91–99. [Google Scholar]
- Gomes, C.M. Protein misfolding in disease and small molecule therapies. Curr. Top. Med. Chem 2012, 12, 2460–2469. [Google Scholar]
- Rodrigues, J.V.; Henriques, B.J.; Lucas, T.G.; Gomes, C.M. Cofactors and metabolites as protein folding helpers in metabolic diseases. Curr. Top. Med. Chem 2012, 12, 2546–2559. [Google Scholar]
- Lucas, T.G.; Henriques, B.J.; Rodrigues, J.V.; Bross, P.; Gregersen, N.; Gomes, C.M. Cofactors and metabolites as potential stabilizers of mitochondrial acyl-coa dehydrogenases. Biochim. Biophys. Acta 2011, 1812, 1658–1663. [Google Scholar]
- Leandro, P.; Gomes, C.M. Protein misfolding in conformational disorders: Rescue of folding defects and chemical chaperoning. Mini Rev. Med. Chem 2008, 8, 901–911. [Google Scholar]
- Leal, S.S.; Botelho, H.M.; Gomes, C.M. Metal ions as modulators of protein conformation and misfolding in neurodegeneration. Coord. Chem. Rev 2012, 256, 2253–2270. [Google Scholar]
- Klunk, W.E.; Xu, C.J.; McClure, R.J.; Panchalingam, K.; Stanley, J.A.; Pettegrew, J.W. Aggregation of beta-amyloid peptide is promoted by membrane phospholipid metabolites elevated in Alzheimer’s disease brain. J. Neurochem 1997, 69, 266–272. [Google Scholar]
- Kim, H.Y.; Kim, Y.; Han, G.; Kim, D.J. Regulation of in vitro abeta1–40 aggregation mediated by small molecules. J. Alzheimer’s Dis 2010, 22, 73–85. [Google Scholar]
- Kawasaki, T.; Kamijo, S. Inhibition of aggregation of amyloid beta42 by arginine-containing small compounds. Biosci. Biotechnol. Biochem 2012, 76, 762–766. [Google Scholar]
- Fung, J.; Darabie, A.A.; McLaurin, J. Contribution of simple saccharides to the stabilization of amyloid structure. Biochem. Biophys. Res. Commun 2005, 328, 1067–1072. [Google Scholar]
- Murakami, G.; Inoue, H.; Tsukita, K.; Asai, Y.; Amagai, Y.; Aiba, K.; Shimogawa, H.; Uesugi, M.; Nakatsuji, N.; Takahashi, R. Chemical library screening identifies a small molecule that downregulates sod1 transcription for drugs to treat amyotrophic lateral sclerosis. J. Biomol. Screen 2011, 16, 405–414. [Google Scholar]
- Limpert, A.S.; Mattmann, M.E.; Cosford, N.D. Recent progress in the discovery of small molecules for the treatment of amyotrophic lateral sclerosis (ALS). Beilstein J. Org. Chem 2013, 9, 717–732. [Google Scholar]
- Ray, S.S.; Nowak, R.J.; Brown, R.H., Jr; Lansbury, P.T., Jr. Small-molecule-mediated stabilization of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants against unfolding and aggregation. Proc. Natl. Acad. Sci. USA 2005, 102, 3639–3644. [Google Scholar]
- Wittung-Stafshede, P. Protein folding inside the cell. Biophys. J 2011, 101, 265–266. [Google Scholar]
- Parone, P.A.; da Cruz, S.; Han, J.S.; McAlonis-Downes, M.; Vetto, A.P.; Lee, S.K.; Tseng, E.; Cleveland, D.W. Enhancing mitochondrial calcium buffering capacity reduces aggregation of misfolded SOD1 and motor neuron cell death without extending survival in mouse models of inherited amyotrophic lateral sclerosis. J. Neurosci 2013, 33, 4657–4671. [Google Scholar]
- Tradewell, M.L.; Cooper, L.A.; Minotti, S.; Durham, H.D. Calcium dysregulation, mitochondrial pathology and protein aggregation in a culture model of amyotrophic lateral sclerosis: Mechanistic relationship and differential sensitivity to intervention. Neurobiol. Dis 2011, 42, 265–275. [Google Scholar]
- Leal, S.S.; Cardoso, I.; Valentine, J.S.; Gomes, C.M. Calcium ions promote superoxide dismutase 1 (SOD1) aggregation into non fibrillar amyloid: A link to toxic effects of calcium overload in amyotrophic lateral sclerosis (ALS)? J. Biol. Chem 2013, 288, 25219–25228. [Google Scholar]
- Furukawa, Y.; Kaneko, K.; Yamanaka, K.; O’Halloran, T.V.; Nukina, N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem 2008, 283, 24167–24176. [Google Scholar]
- Furukawa, Y.; O’Halloran, T.V. Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo- and reduced form of SOD1, leading to unfolding and oxidative aggregation. J. Biol. Chem 2005, 280, 17266–17274. [Google Scholar]
- Jonsson, P.A.; Graffmo, K.S.; Andersen, P.M.; Brannstrom, T.; Lindberg, M.; Oliveberg, M.; Marklund, S.L. Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain 2006, 129, 451–464. [Google Scholar]
- Lelie, H.L.; Liba, A.; Bourassa, M.W.; Chattopadhyay, M.; Chan, P.K.; Gralla, E.B.; Miller, L.M.; Borchelt, D.R.; Valentine, J.S.; Whitelegge, J.P. Copper and zinc metallation status of copper-zinc superoxide dismutase from amyotrophic lateral sclerosis transgenic mice. J. Biol. Chem 2011, 286, 2795–2806. [Google Scholar]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. Hmdb 3.0—The human metabolome database in 2013. Nucleic Acids Res 2013, 41, D801–D807. [Google Scholar]
- Street, T.O.; Bolen, D.W.; Rose, G.D. A molecular mechanism for osmolyte-induced protein stability. Proc. Natl. Acad. Sci. USA 2006, 103, 13997–14002. [Google Scholar]
- Durazo, A.; Shaw, B.F.; Chattopadhyay, M.; Faull, K.F.; Nersissian, A.M.; Valentine, J.S.; Whitelegge, J.P. Metal-free superoxide dismutase-1 and three different amyotrophic lateral sclerosis variants share a similar partially unfolded beta-barrel at physiological temperature. J. Biol. Chem 2009, 284, 34382–34389. [Google Scholar]
- Hawe, A.; Sutter, M.; Jiskoot, W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res 2008, 25, 1487–1499. [Google Scholar]
- Tiwari, A.; Xu, Z.; Hayward, L.J. Aberrantly increased hydrophobicity shared by mutants of Cu,Zn-superoxide dismutase in familial amyotrophic lateral sclerosis. J. Biol. Chem 2005, 280, 29771–29779. [Google Scholar]
- Munch, C.; Bertolotti, A. Exposure of hydrophobic surfaces initiates aggregation of diverse ALS-causing superoxide dismutase-1 mutants. J. Mol. Biol 2010, 399, 512–525. [Google Scholar]
- LeVine, H., 3rd. Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999, 309, 274–284. [Google Scholar]
- LeVine, H., 3rd. Thioflavine T interaction with synthetic Alzheimer’s disease beta-amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993, 2, 404–410. [Google Scholar]
- Biancalana, M.; Koide, S. Molecular mechanism of thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta 2010, 1804, 1405–1412. [Google Scholar]
- Jan, A.; Gokce, O.; Luthi-Carter, R.; Lashuel, H.A. The ratio of monomeric to aggregated forms of abeta40 and abeta42 is an important determinant of amyloid-beta aggregation, fibrillogenesis, and toxicity. J. Biol. Chem 2008, 283, 28176–28189. [Google Scholar]
- Maezawa, I.; Hong, H.S.; Liu, R.; Wu, C.Y.; Cheng, R.H.; Kung, M.P.; Kung, H.F.; Lam, K.S.; Oddo, S.; Laferla, F.M.; et al. Congo red and thioflavin-t analogs detect abeta oligomers. J. Neurochem 2008, 104, 457–468. [Google Scholar]
- Pedersen, J.S.; Dikov, D.; Flink, J.L.; Hjuler, H.A.; Christiansen, G.; Otzen, D.E. The changing face of glucagon fibrillation: Structural polymorphism and conformational imprinting. J. Mol. Biol 2006, 355, 501–523. [Google Scholar]
- Sandelin, E.; Nordlund, A.; Andersen, P.M.; Marklund, S.S.; Oliveberg, M. Amyotrophic lateral sclerosis-associated copper/zinc superoxide dismutase mutations preferentially reduce the repulsive charge of the proteins. J. Biol. Chem 2007, 282, 21230–21236. [Google Scholar]
- Das, U.; Hariprasad, G.; Ethayathulla, A.S.; Manral, P.; Das, T.K.; Pasha, S.; Mann, A.; Ganguli, M.; Verma, A.K.; Bhat, R.; et al. Inhibition of protein aggregation: Supramolecular assemblies of arginine hold the key. PLoS One 2007, 2, e1176. [Google Scholar]
- Tsumoto, K.; Ejima, D.; Kita, Y.; Arakawa, T. Review: Why is arginine effective in suppressing aggregation? Protein Pept. Lett 2005, 12, 613–619. [Google Scholar]
- Shah, D.; Shaikh, A.R.; Peng, X.; Rajagopalan, R. Effects of arginine on heat-induced aggregation of concentrated protein solutions. Biotechnol. Prog 2011, 27, 513–520. [Google Scholar]
- Chattopadhyay, M.; Durazo, A.; Sohn, S.H.; Strong, C.D.; Gralla, E.B.; Whitelegge, J.P.; Valentine, J.S. Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl. Acad. Sci. USA 2008, 105, 18663–18668. [Google Scholar]
- Oztug Durer, Z.A.; Cohlberg, J.A.; Dinh, P.; Padua, S.; Ehrenclou, K.; Downes, S.; Tan, J.K.; Nakano, Y.; Bowman, C.J.; Hoskins, J.L.; et al. Loss of metal ions, disulfide reduction and mutations related to familial als promote formation of amyloid-like aggregates from superoxide dismutase. PLoS One 2009, 4, e5004. [Google Scholar]
- Filipe, V.; Hawe, A.; Jiskoot, W. Critical evaluation of nanoparticle tracking analysis (NTA) by nanosight for the measurement of nanoparticles and protein aggregates. Pharm. Res 2010, 27, 796–810. [Google Scholar]
- Elam, J.; Taylor, A.; Strange, R.; Antonyuk, S.; Doucette, P.; Rodriguez, J.; Hasnain, S.; Hayward, L.; Valentine, J.; Yeates, T.; et al. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat. Struct. Biol 2003, 10, 461–467. [Google Scholar]
- Chan, P.K.; Chattopadhyay, M.; Sharma, S.; Souda, P.; Gralla, E.B.; Borchelt, D.R.; Whitelegge, J.P.; Valentine, J.S. Structural similarity of wild-type and ALS-mutant superoxide dismutase-1 fibrils using limited proteolysis and atomic force microscopy. Proc. Natl. Acad. Sci. USA 2013, 110, 10934–10939. [Google Scholar]
- Ahl, I.M.; Lindberg, M.J.; Tibell, L.A. Coexpression of yeast copper chaperone (YCCS) and Cu/Zn-superoxide dismutases in Escherichia coli yields protein with high copper contents. Protein Expression Purif 2004, 37, 311–319. [Google Scholar]
- Potter, S.Z.; Zhu, H.; Shaw, B.F.; Rodriguez, J.A.; Doucette, P.A.; Sohn, S.H.; Durazo, A.; Faull, K.F.; Gralla, E.B.; Nersissian, A.M.; et al. Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J. Am. Chem. Soc 2007, 129, 4575–4583. [Google Scholar]
- Sabel, C.E.; Neureuther, J.M.; Siemann, S. A spectrophotometric method for the determination of zinc, copper, and cobalt ions in metalloproteins using zincon. Anal. Biochem 2010, 397, 218–226. [Google Scholar]

| Compound | Compound properties | CSF levels (μM) | Sporadic ALS | ||
|---|---|---|---|---|---|
| Type | Physiological charge | Normal | Altered | ||
| l-Arginine | +1 | 15–25 | 6–21 | decreased | |
| l-Lysine | amino acid | +1 | 18–30 | 45 | decreased |
| l-Ornithine | +1 | 4–8 | 7–8 | unchanged | |
| Glyceric acid | sugar acid | −1 | 34 | n.a. | decreased |
| Phthalate | exogenous | −2 | n.a. | n.a | unchanged |
| Fructose | monosaccharide | 0 | 160–240 | n.a. | decreased |
| Erythronic Acid | −1 | 5 | n.a | unchanged | |
| Compound | Lag time tlag, h | Aggregation rate kapp, h−1 |
|---|---|---|
| Control (no compound) | 88 | 0.040 |
| Erythronic Acid | 47 | 0.093 |
| l-Ornithine | 20 | 0.062 |
| Phthalate | 24 | 0.059 |
| l-Arginine | 41 | 0.054 |
| Glyceric acid | 39 | 0.044 |
| Fructose | 17 | 0.044 |
| l-Lysine | n.d. | 0.019 |
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Cristóvão, J.S.; Leal, S.S.; Cardoso, I.; Gomes, C.M. Small Molecules Present in the Cerebrospinal Fluid Metabolome Influence Superoxide Dismutase 1 Aggregation. Int. J. Mol. Sci. 2013, 14, 19128-19145. https://doi.org/10.3390/ijms140919128
Cristóvão JS, Leal SS, Cardoso I, Gomes CM. Small Molecules Present in the Cerebrospinal Fluid Metabolome Influence Superoxide Dismutase 1 Aggregation. International Journal of Molecular Sciences. 2013; 14(9):19128-19145. https://doi.org/10.3390/ijms140919128
Chicago/Turabian StyleCristóvão, Joana S., Sónia S. Leal, Isabel Cardoso, and Cláudio M. Gomes. 2013. "Small Molecules Present in the Cerebrospinal Fluid Metabolome Influence Superoxide Dismutase 1 Aggregation" International Journal of Molecular Sciences 14, no. 9: 19128-19145. https://doi.org/10.3390/ijms140919128






