Discovery and in Vivo Evaluation of Novel RGD-Modified Lipid-Polymer Hybrid Nanoparticles for Targeted Drug Delivery
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Curcumin (Cur)–Lipid-Shell and Polymer Core Nanoparticles (lpNPs)
| Drug/Polymer (%, w/w) | Size (nm) | Polydispersity Index (PDI) | DL (%) | EE (%) |
|---|---|---|---|---|
| 2 | 201.9 ± 7.8 | 0.195 ± 0.05 | 2.0 ± 0.01 | 100 ± 0.24 |
| 3 | 206.36 ± 6.1 | 0.192 ± 0.07 | 2.96 ± 0.02 | 98.67 ± 0.67 |
| 4 | 202.4 ± 5.2 | 0.202 ± 0.03 | 3.91 ± 0.02 | 97.75 ± 0.5 |
| 5 | 216.6 ± 4.7 | 0.205 ± 0.02 | 4.80 ± 0.03 | 96.0 ± 0.6 |
| 7 | 260.5 ± 8.1 | 0.282 ± 0.07 | 4.95 ± 0.07 | 70.71 ± 1.0 |
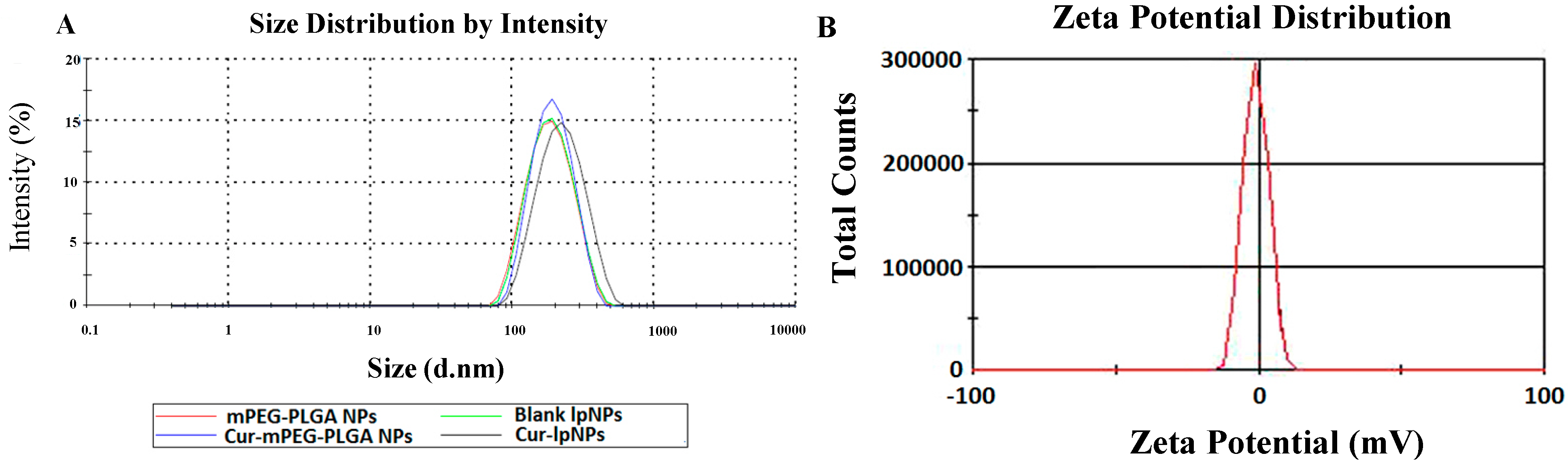

2.2. In Vitro Cytotoxicity Evaluation

2.3. In Vitro Apoptosis Induction Effect

2.4. Cellular Uptake of Cur–lpNPs by Flow Cytometry Analysis
2.5. In Vivo Anti-Tumor Effect in Subcutaneous Tumor Model


2.6. Determination of Apoptosis
2.7. Quantitative Assessment of Microvessel Density (MVD)


3. Experimental Section
3.1. Materials, Cell Lines and Animals
3.2. Preparation and Characterization of Cur–lpNPs
3.3. In Vitro Cytotoxicity
3.4. Cellular Apoptosis Assay
3.5. Cellular Uptake of Cur–lpNPS by Flow Cytometry Analysis
3.6. Subcutaneous Tumor Mouse Model
3.7. Detection of Apoptosis
3.8. Quantitative Assessment of MVD
3.9. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Singh, S. From exotic spice to modern drug? Cell 2007, 130, 765–768. [Google Scholar] [CrossRef]
- Jagetia, G.C.; Aggarwal, B.B. Spicing up of the immune system by curcumin. J. Clin. Immunol. 2007, 27, 19–35. [Google Scholar]
- Tang, H.; Murphy, C.J.; Zhang, B.; Shen, Y.; van Kirk, E.A.; Murdoch, W.J.; Radosz, M. Curcumin polymers as anticancer conjugates. Biomaterials 2010, 31, 7139–7149. [Google Scholar]
- Gong, C.Y.; Wang, C.; Wang, Y.J.; Wu, Q.J.; Zhang, D.D.; Luo, F.; Qian, Z.Y. Efficient inhibition of colorectal peritoneal carcinomatosis by drug loaded micelles in thermosensitive hydrogel composites. Nanoscale 2012, 4, 3095–3104. [Google Scholar]
- Yallapu, M.M.; Jaggi, M.; Chauhan, S.C. Curcumin nanoformulations: A future nanomedicine for cancer. Drug Discov. Today 2012, 17, 71–80. [Google Scholar]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar]
- Liu, L.; Sun, L.; Wu, Q.J.; Guo, W.H.; Li, L.; Chen, Y.S.; Li, Y.C.; Gong, C.Y.; Qian, Z.Y.; Wei, Y.Q. Curcumin loaded polymeric micelles inhibit breast tumor growth and spontaneous pulmonary metastasis. Int. J. Pharm. 2013, 443, 175–182. [Google Scholar]
- Hadinoto, K.; Sundaresan, A.; Cheow, W.S. Lipid-polymer hybrid nanoparticles as a new generation therapeutic delivery platform: A review. Eur. J. Pharm. Biopharm. 2013, 85, 427–443. [Google Scholar]
- Zhao, P.; Wang, H.; Yu, M.; Liao, Z.; Wang, X.; Zhang, F.; Ji, W.; Wu, B.; Han, J.; Zhang, H. Paclitaxel loaded folic acid targeted nanoparticles of mixed lipid-shell and polymer-core: In vitro and in vivo evaluation. Eur. J. Pharm. Biopharm. 2012, 81, 248–256. [Google Scholar]
- Chitkara, D.; Kumar, N. BSA-PLGA-Based core-shell nanoparticles as carrier system for water-soluble drugs. Pharm. Res. 2013, 30, 2396–2409. [Google Scholar] [CrossRef]
- Liu, Y.; Metasebya, S.; Samuel, A. Perspectives and potential applications of nanomedicine in breast and prostate cancer. Med. Res. Rev. 2013, 33, 3–32. [Google Scholar] [CrossRef]
- Patrick, V.; Michel, K. Medicinal chemistry based approaches and nanotechnology-based systems to improve CNS drug targeting and delivery. Med. Res. Rev. 2013, 33, 457–516. [Google Scholar]
- Jagat, R.K.; Sishir, K.K.; Rupinder, K.K. Survivin signaling in clinical oncology: A multifaceted dragon. Med. Res. Rev. 2013, 33, 765–789. [Google Scholar]
- Chan, J.M.; Zhang, L.F.; Yuet, K.P.; Liao, G.; Rhee, J.W.; Langer, R.; Farokhzad, O.C. PLGA–lecithin–PEG core-shell nanoparticles for controlled drug delivery. Biomaterials 2009, 30, 1627–1634. [Google Scholar]
- Li, J.; He, Z.; Yu, S.; Li, S.; Ma, Q.; Yu, Y.; Zhang, J.; Li, R.; Zheng, Y.; He, G.; et al. Micelles based on methoxy poly(ethylene glycol)-cholesterol conjugate for controlled and targeted drug delivery of a poorly water soluble drug. J. Biomed. Nanotechnol. 2012, 8, 809–817. [Google Scholar]
- Messerschmidt, S.K.E.; Musyanovych, A.; Altvater, M.; Scheurich, P.; Pfizenmaier, K.; Landfester, K.; Kontermann, R.E. Targeted lipid-coated nanoparticles: Delivery of tumor necrosis factor-functionalized articles to tumor cells. J. Control. Release 2009, 137, 69–72. [Google Scholar]
- Fang, D.-L.; Chen, Y.; Xu, B.; Ren, K.; He, Z.-Y.; He, L.-L.; Lei, Y.; Fan, C.-M.; Song, X.-R. Development of lipid-shell and polymer core nanoparticles with water-soluble salidroside for anti-cancer therapy. Int. J. Mol. Sci. 2014, 15, 3373–3388. [Google Scholar]
- Jeanne, L.; Nathalie, M.; Lucie, L.; Céline, L.; Michel, B.; Daniel, S.; Jean, H. Design, synthesis and evaluation of enhanced DNA binding new lipopolythioureas. Bioconj. Chem. 2006, 17, 1200–1208. [Google Scholar]
- Zeng, S.; Wu, F.B.; Li, B.; Song, X.R.; Zheng, Y.; He, G.; Peng, C.; Huang, W. Synthesis, characterization and evaluation of a novel amphiphilic polymer RGD–PEG–Chol for target drug delivery system. Sci. World J. 2014. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Lin, D.; Wu, F.; Guo, L.; He, G.; Ouyang, L.; Song, X.; Huang, W.; Li, X. Discovery and in Vivo Evaluation of Novel RGD-Modified Lipid-Polymer Hybrid Nanoparticles for Targeted Drug Delivery. Int. J. Mol. Sci. 2014, 15, 17565-17576. https://doi.org/10.3390/ijms151017565
Zhao Y, Lin D, Wu F, Guo L, He G, Ouyang L, Song X, Huang W, Li X. Discovery and in Vivo Evaluation of Novel RGD-Modified Lipid-Polymer Hybrid Nanoparticles for Targeted Drug Delivery. International Journal of Molecular Sciences. 2014; 15(10):17565-17576. https://doi.org/10.3390/ijms151017565
Chicago/Turabian StyleZhao, Yinbo, Dayong Lin, Fengbo Wu, Li Guo, Gu He, Liang Ouyang, Xiangrong Song, Wei Huang, and Xiang Li. 2014. "Discovery and in Vivo Evaluation of Novel RGD-Modified Lipid-Polymer Hybrid Nanoparticles for Targeted Drug Delivery" International Journal of Molecular Sciences 15, no. 10: 17565-17576. https://doi.org/10.3390/ijms151017565





