Sterols from Mytilidae Show Anti-Aging and Neuroprotective Effects via Anti-Oxidative Activity
Abstract
:1. Introduction
2. Results and Discussion
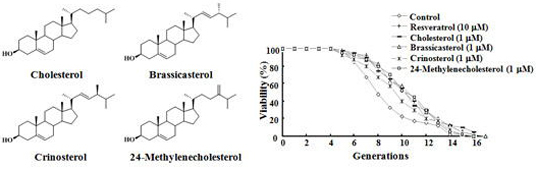
2.1. SF, Sterols, and Their Anti-Aging Activities

2.2. CHOL Improved the Oxidative Resistance of Yeast

2.3. CHOL Decreased ROS and MDA Production



2.4. Anti-Aging Effects of CHOL Diminished in Uth1, Skn7, Sod1, and Sod2 Mutations with a K6001 Background
2.5. SF and CHOL Exerted Neuroprotective Effects on PC12 Cells
3. Experimental Section
3.1. Extraction and Isolation
3.2. Chemical Analysis
3.3. Strains, Culture Media, and Lifespan Assay
3.4. Anti-Oxidative Stress Assay
3.5. ROS and MDA Assays
3.6. RT-PCR Assay
3.7. Bioassay of NGF Mimic Activity and Neuroprotection Assay on PC12 Cells
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Abbreviations
| AD | Alzheimer’s disease |
| CHOL | cholesterol |
| DMEM | Dulbecco’s modified eagle medium |
| HPLC | high performance liquid chromatography |
| MDA | malonaldehyde |
| NGF | nerve growth factor |
| PBS | phosphate buffer solution |
| ROS | reactive oxidative species |
| SF | sterol fraction |
| YPD | yeast peptone d-glucose |
Conflicts of Interest
References
- Blagosklonny, M.V. Validation of anti-aging drugs by treating age-related diseases. Aging-US 2009, 1, 281–288. [Google Scholar]
- Walhovd, K.B.; Fjell, A.M.; Espeseth, T. Cognitive decline and brain pathology in aging-need for a dimensional, lifespan and systems vulnerability view. Scand. J. Psychol. 2014, 55, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wang, X.; Xiang, Q.; Meng, X.; Peng, Y.; Du, N.; Liu, Z.; Sun, Q.; Wang, C.; Liu, X. Astaxanthin alleviates brain aging in rats by attenuating oxidative stress and increasing BDNF levels. Food Funct. 2014, 5, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.H.; Park, S.W.; Brooks, N.; Lang, B.T.; Vemuganti, R. PPARγ agonist rosiglitazone is neuroprotective after traumatic brain injury via anti-inflammatory and anti-oxidative mechanisms. Brain Res. 2008, 1244, 164–172. [Google Scholar]
- Chen, W.; Qi, J.; Feng, F.; Wang, M.D.; Bao, G.; Wang, T.; Xiang, M.; Xie, W.F. Neuroprotective effect of allicin against traumatic brain injury via Akt/endothelial nitric oxide synthase pathway-mediated anti-inflammatory and anti-oxidative activities. Neurochem. Int. 2014, 68, 28–37. [Google Scholar] [CrossRef]
- Kim, D.; Nguyen, M.D.; Dobbin, M.M.; Fischer, A.; Sananbenesi, F.; Rodgers, J.T.; Delalle, I.; Baur, J.A.; Sui, G.; Armour, S.M.; et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. Embo J. 2007, 26, 3169–3179. [Google Scholar] [CrossRef]
- Thapa, A.; Vernon, B.C.; de la Peña, K.; Soliz, G.; Moreno, H.A.; López, G.P.; Chi, E.Y. Membrane-mediated neuroprotection by curcumin from Amyloid-β-peptide-induced toxicity. Langmuir 2013, 29, 11713–11723. [Google Scholar] [CrossRef] [PubMed]
- Hamaguchi, T.; Ono, K.; Yamada, M. Curcumin and Alzheimer’s disease. CNS Neurosci. Ther. 2010, 16, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Yu, J.T.; Zhu, X.C.; Wang, H.F.; Tan, M.S.; Cao, L.; Zhang, Q.Q.; Gao, L.; Shi, J.Q.; Zhang, Y.D.; et al. Acute metformin preconditioning confers neuroprotection against focal cerebral ischaemia by pre-activation of AMPK-dependent autophagy. Br. J. Pharmacol. 2014, 171, 3146–3157. [Google Scholar] [CrossRef]
- Kickstein, E.; Krauss, S.; Thornhill, P.; Rutschow, D.; Zeller, R.; Sharkey, J.; Williamson, R.; Fuchs, M.; Köhler, A.; Glossmann, H.; et al. Biguanide metformin acts on tau phosphorylation via mTOR/protein phosphatase 2A (PP2A) signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 21830–21835. [Google Scholar] [CrossRef]
- Pan, T.H.; Kondo, S.; Zhu, W.; Xie, W.; Jankovic, J.; Le, W. Neuroprotection of rapamycin in lactacystin-induced neurodegeneration via autophagy enhancement. Neurobiol. Dis. 2008, 32, 16–25. [Google Scholar] [PubMed]
- Orr, M.E.; Salinas, A.; Buffenstein, R.; Oddo, S. Mammalian target of rapamycin hyperactivity mediates the detrimental effects of a high sucrose diet on Alzheimer’s disease pathology. Neurobiol. Aging 2014, 35, 1233–1242. [Google Scholar] [CrossRef] [PubMed]
- Thies, W.; Bleiler, L.; Alzheimer’s, A. 2013 Alzheimer’s disease facts and figures. Alzheimers Dement. 2013, 9, 208–245. [Google Scholar] [CrossRef] [PubMed]
- Prince, M.; Prina, M.; Guerchet, M. World Alzheimer Report 2013; Alzheimer’s Disease International (ADI): London, UK, September 2013. [Google Scholar]
- Weng, Y.; Xiang, L.; Matsuura, A.; Zhang, Y.; Huang, Q.; Qi, J. Ganodermasides A and B, two novel anti-aging ergosterols from spores of a medicinal mushroom Ganoderma lucidum on yeast via UTH1 gene. Bioorg. Med. Chem. 2010, 18, 999–1002. [Google Scholar] [CrossRef]
- Xiang, L.; Sun, K.; Lu, J.; Weng, Y.; Taoka, A.; Sakagami, Y.; Qi, J. Anti-aging effects of phloridzin, an apple polyphenol, on yeast via the SOD and Sir2 genes. Biosci. Biotechnol. Biochem. 2011, 75, 854–858. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Cao, S.; Pei, L.; Matsuura, A.; Xiang, L.; Qi, J. A steroidal saponin from Ophiopogon japonicus extends the lifespan of yeast via the pathway involved in SOD and UTH1. Int. J. Mol. Sci. 2013, 14, 4461–4475. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Wang, H.; Zheng, G.; Zhang, R. Studies on chemical constituents of Meretrix meretrix. Chin. J. Mar. Drugs 2003, 22, 31–32. [Google Scholar]
- Bergmann, W.; Feeney, R.J. Contributions to the study of marine products. XXIII. Sterols from sponges of the family Haliclonidae. J. Org. Chem. 1949, 14, 1078–1084. [Google Scholar] [CrossRef]
- Lang, R.W.; Djerassi, C. Stereochemical aspects of acid-catalyzed cyclopropane ring-opening reactions. A stereospecific pathway to crinosterol and brassicasterol. Helv. Chim. Acta 1982, 65, 407–418. [Google Scholar] [CrossRef]
- Kiyoshi, S.; Kyosuke, T. Steroids. XXX. The synthesis of brassicasterol. Chem. Pharm. Bull. 1963, 11, 650–653. [Google Scholar] [CrossRef]
- Bolker, H.I. Crinosterol: A unique sterol from a comatulid crinoids. Nature 1967, 213, 905–906. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zhang, C.; Zeng, L.; Su, J. Synthesis of polyhydroxysterols (V): Efficient and stereospecific synthesis of 24-methylene-cholest-5-ene-3β, 7α-diol and its C-7 epimer. Steroids 2004, 69, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.A.; Reynolds, R.M. Evolutionary and mechanistic theories of aging. Annu. Rev. Entomol. 2005, 50, 421–445. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.L. Another cholesterol hypothesis: Cholesterol as antioxidant. Free Radic. Biol. Med. 1991, 11, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Olsher, M.; Yoon, S.I.; Chong, P.L. Role of sterol superlattice in free radical-induced sterol oxidation in lipid membranes. Biochemistry 2005, 44, 2080–2087. [Google Scholar] [CrossRef] [PubMed]
- Raha, S.; Robinson, B.H. Mitochondria, oxygen free radicals, disease and ageing. Trends Biochem. Sci. 2000, 25, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Turton, H.E.; Dawes, I.W.; Grant, C.M. Saccharomyces cerevisiae exhibits a yAP-1-mediated adaptive response to malondialdehyde. J. Bacteriol. 1997, 179, 1096–1101. [Google Scholar] [PubMed]
- Bandara, P.D.S.; Flattery-O’Brien, J.A.; Grant, C.M.; Dawes, I.W. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Curr. Genet. 1998, 34, 259–268. [Google Scholar] [CrossRef]
- Mulford, K.E.; Fassler, J.S. Association of the Skn7 and Yap1 transcription factors in the Saccharomyces cerevisiae oxidative stress response. Eukaryot. Cell 2011, 10, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Roberti, G.; Mantelli, F.; Macchi, I.; Massaro-Giordano, M.; Centofanti, M. Nerve growth factor modulation of retinal ganglion cell physiology. J. Cell. Physiol. 2014, 229, 1130–1133. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Luo, Y.; Tang, R.; Zhang, H.; Ye, Y.; Xiang, L.; Qi, J. Neuritogenic monoglyceride derived from the constituent of a marine fish for activating the PI3K/ERK/CREB signaling pathways in PC12 cells. Int. J. Mol. Sci. 2013, 14, 24200–24210. [Google Scholar] [CrossRef] [PubMed]
- SPSS Biostatistics Software, version 16.0; SPSS Inc.: Chicago, IL, USA, 2008.
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Lin, Y.; Cao, X.; Xiang, L.; Qi, J. Sterols from Mytilidae Show Anti-Aging and Neuroprotective Effects via Anti-Oxidative Activity. Int. J. Mol. Sci. 2014, 15, 21660-21673. https://doi.org/10.3390/ijms151221660
Sun Y, Lin Y, Cao X, Xiang L, Qi J. Sterols from Mytilidae Show Anti-Aging and Neuroprotective Effects via Anti-Oxidative Activity. International Journal of Molecular Sciences. 2014; 15(12):21660-21673. https://doi.org/10.3390/ijms151221660
Chicago/Turabian StyleSun, Yujuan, Yanfei Lin, Xueli Cao, Lan Xiang, and Jianhua Qi. 2014. "Sterols from Mytilidae Show Anti-Aging and Neuroprotective Effects via Anti-Oxidative Activity" International Journal of Molecular Sciences 15, no. 12: 21660-21673. https://doi.org/10.3390/ijms151221660