Sll0528, a Site-2-Protease, Is Critically Involved in Cold, Salt and Hyperosmotic Stress Acclimation of Cyanobacterium Synechocystis sp. PCC 6803
Abstract
:1. Introduction
2. Results and Discussion
2.1. Expression Profile of S2Ps under Different Conditions

2.2. Construction of the sll0528 Knockout Mutant

2.3. Varied Phenotype of sll0528 Mutant under Different Conditions
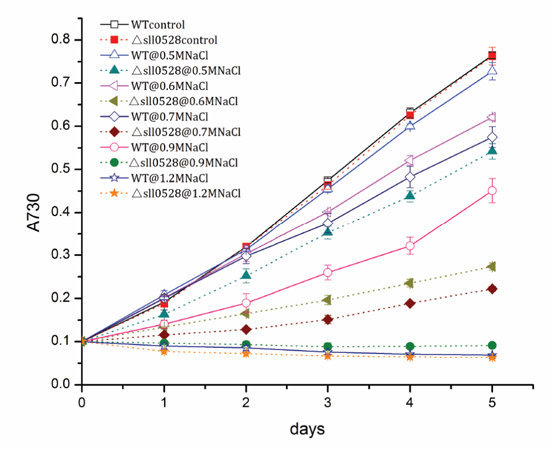
2.3.1. Sll0528 Is Crucial for Acclimation to Salt Stress
2.3.2. Sll0528 Is Indispensable for Acclimation to Hyperosmotic Stress


2.3.3. Sll0528 Is Crucial for Cold Acclimation

2.3.4. Sll0528 Is Dispensable for Acclimation to High Light and Mixotrophic Growth

2.4. Recombinant Sll0528 Has Proteolytic Activity
3. Experimental Section
3.1. Strains, Culture and Stress Conditions

3.2. RNA Isolation and Quantitative RT-PCR
3.3. Construction of sll0528 Mutant
3.4. Measurement of Physiological Parameters
3.5. Expression of Recombinant Sll0528 and in Vitro Proteolytic Activity Assay
3.6. Data Analysis
| Primer Name | Sequence of Primer (5'–3') |
|---|---|
| sll0528-L | GGAAGCCTTTACTGCTGAAGAT |
| sll0528-R | TGTCGGCACCAATAACCAAG |
| sll0862-L | GGCAAATGCGGGAAGAAG |
| sll0862-R | TGTCACCGAGCACAGTGGT |
| slr0643-L | GGTTTGTCCACTGCTCTACT |
| slr0643-R | GGCTGTGATGATTTCTGC |
| slr1821-L | TTGGATGGTGGGCAATTG |
| slr1821-R | ATACCCCTAAGCTCAGCAGAAG |
| rnpB-L | CCACTGAAAAGGGTAAGGG |
| rnpB-R | CTCCGACCTTGCTTCCA |
| P1 | CGAGCTCCTACGGAAGACATCAAACACG |
| P2 | CCGCTCGAGGGCGAAATGTTGACCTTGAC |
| P3 | GGAATTCCATATGCTGGTTATGGTGGTTTACTGA |
| P4 | CGCGGATCCATGTTAAGAATTGCCTGAGTG |
| P5 | CGCGGATCCTCATCAGTGCCAACATAG |
| P6 | CGAGCTCGGTAAACCAGCAATAGACAT |
| P7 | CAAGTACAC CTAACAGTTGA |
| P8 | ATGTTAAGCCTCAGTTTAG |
| P9 | CGCGGATCCATGTTAAGCCTCAGTTTAGGGG |
| P10 | CCGGAATTCCTAGGCGGCGGAGGTTTGCAG |
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guerrero, F.; Carbonell, V.; Cossu, M.; Correddu, D.; Jones, P.R. Ethylene synthesis and regulated expression of recombinant protein in Synechocystis sp. PCC 6803. PLoS One 2012, 7, e50470. [Google Scholar]
- Liu, X.; Sheng, J.; Curtiss, R., III. Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 6899–6904. [Google Scholar]
- Oliver, J.W.; Machado, I.M.; Yoneda, H.; Atsumi, S. Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. Proc. Natl. Acad. Sci. USA 2013, 110, 1249–1254. [Google Scholar]
- Los, D.A.; Zorina, A.; Sinetova, M.; Kryazhov, S.; Mironov, K.; Zinchenko, V.V. Stress sensors and signal transducers in cyanobacteria. Sensors 2010, 10, 2386–2415. [Google Scholar]
- Chen, G.; Zhang, X. New insights into S2P signaling cascades: Regulation, variation, and conservation. Pro. Sci. 2010, 19, 2015–2030. [Google Scholar]
- Brown, M.S.; Goldstein, J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 1997, 89, 331–340. [Google Scholar]
- Brown, M.S.; Goldstein, J.L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 1999, 96, 11041–11048. [Google Scholar]
- Brown, M.S.; Ye, J.; Rawson, R.B.; Goldstein, J.L. Regulated intramembrane proteolysis: Review a control mechanism conserved from bacteria to humans. Cell 2000, 100, 391–398. [Google Scholar]
- Chen, G.; Bi, Y.R.; Li, N. EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. Cell Mol. Biol. 2005, 41, 364–375. [Google Scholar]
- Chen, G.; Law, K.; Ho, P.; Zhang, X.; Li, N. EGY2, a chloroplast membrane metalloprotease, plays a role in hypocotyl elongation in Arabidopsis. Mol. Biol. Rep. 2012, 39, 2147–2155. [Google Scholar]
- Chen, G.; Wen, P.P.; Qin, C.; Wang, Y. Expression of green fluorescent protein-tagged S2P homologs in Synechocystis sp. PCC 6803. J. South China Uni. Technol. 2012, 40, 122–127. [Google Scholar]
- Zhang, X.; Chen, G.; Qin, C.; Wang, Y.; Wei, D. Slr0643, an S2P homologue, is essential for acid acclimation in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 2012, 158, 2765–2780. [Google Scholar]
- Guo, D.; Gao, X.R.; Li, H.; Zhang, T.; Chen, G.; Huang, P.B. EGY1 plays a role in regulation of endodermal plastid size and number that are involved in ethylene-dependent gravitropism of light-grown Arabidopsis hypocotyls. Plant Mol. Biol. 2008, 66, 345–360. [Google Scholar]
- Chen, K.; Gu, L.; Xiang, X.; Lynch, M.; Zhou, R. Identification and characterization of five intramembrane metalloproteases in Anabaena variabilis. J. Bacteriol. 2012, 194, 6105–6115. [Google Scholar]
- Kanesaki, Y.; Suzuki, I.; Allakhverdiev, S.I.; Mikami, K.; Murata, N. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 2002, 290, 339–348. [Google Scholar]
- Marin, K.; Kanesaki, Y.; Los, D.A.; Murata, N.; Suzuki, I.; Hagemann, M. Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant Physiol. 2004, 136, 3290–3300. [Google Scholar]
- Shoumskaya, M.A.; Paithoonrangsarid, K.; Kanesaki, Y.; Los, D.A.; Zinchenko, V.V.; Tanticharoen, M. Identical Hik-Rre systems are involved in perception and transduction of salt signals and hyperosmotic signals but regulate the expression of individual genes to different extents in synechocystis. J. Biol. Chem. 2005, 280, 21531–21538. [Google Scholar]
- Shapiguzov, A.; Lyukevich, A.A.; Allakhverdiev, S.I.; Sergeyenko, T.V.; Suzuki, I.; Murata, N. Osmotic shrinkage of cells of Synechocystis sp. PCC 6803 by water efflux via aquaporins regulates osmostress-inducible gene expression. Microbiology 2005, 151, 447–455. [Google Scholar]
- Paithoonrangsarid, K.; Shoumskaya, M.A.; Kanesaki, Y.; Satoh, S.; Tabata, S.; Los, D.A. Five histidine kinases perceive osmotic stress and regulate distinct sets of genes in Synechocystis. J. Biol. Chem. 2004, 279, 53078–53086. [Google Scholar]
- Inaba, M.; Suzuki, I.; Szalontai, B.; Kanesaki, Y.; Los, D.A.; Hayashi, H. Gene-engineered rigidification of membrane lipids enhances the cold inducibility of gene expression in Synechocystis. J. Biol. Chem. 2003, 278, 12191–12198. [Google Scholar]
- Kanesaki, Y.; Shiwa, Y.; Tajima, N.; Suzuki, M.; Watanabe, S.; Sato, N. Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res. 2012, 19, 67–79. [Google Scholar]
- Danika, T.; Bjorn, V.; Annegret, W.; Salim, A.; Wolfgang, R.H. Microevolution in cyanobacteria: Re-sequencing a motile substrain of Synechocystis sp PCC 6803. DNA Res. 2012, 19, 435–448. [Google Scholar]
- Tajima, N.; Sato, S.; Maruyama, F.; Kaneko, T.; Sasaki, N.V.; Kurokawa, K. Genomic structure of the cyanobacterium Synechocystis sp PCC 6803 strain GT-S. DNA Res. 2011, 18, 393–399. [Google Scholar]
- Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 2011, 35, 87–123. [Google Scholar]
- Jittawuttipoka, T.; Planchon, M.; Spalla, O.; Benzerara, K.; Guyot, F.; Cassier-Chauvat, C. Multidisciplinary evidences that Synechocystis PCC6803 exopolysaccharides operate in cell sedimentation and protection against salt and metal stresses. PLoS One 2013, 8, e55564. [Google Scholar]
- Nikkinen, H.L.; Hakkila, K.; Gunnelius, L.; Huokko, T.; Pollari, M.; Tyystjarvi, T. The SigB σ factor regulates multiple salt acclimation responses of the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2012, 158, 514–523. [Google Scholar]
- Los, D.A.; Ray, M.K.; Murata, N. Differences in the control of the temperature-dependent expression of four genes for desaturases in Synechocystis sp. PCC 6803. Mol. Microbiol. 1997, 25, 1167–1175. [Google Scholar]
- Mironov, K.S.; Sidorov, R.A.; Trofimova, M.S.; Bedbenov, V.S.; Tsydendambaev, V.D.; Allakhverdiev, S.I. Light-dependent cold-induced fatty acid unsaturation, changes in membrane fluidity, and alterations in gene expression in Synechocystis. Biochim. Biophys. Acta 2012, 1817, 1352–1359. [Google Scholar]
- Wada, H.; Murata, N. Temperature-induced changes in the fatty acid composition of the cyanobacterium, Synechocystis PCC6803. Plant Physiol. 1990, 92, 1062–1069. [Google Scholar]
- Suzuki, I.; Kanesaki, Y.; Mikami, K.; Kanehisa, M.; Murata, N. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 2001, 40, 235–244. [Google Scholar]
- Suzuki, I.; Los, D.A.; Kanesaki, Y.; Mikami, K.; Murata, N. The pathway for perception and transduction of low-temperature signals in Synechocystis. EMBO J. 2000, 19, 1327–1334. [Google Scholar]
- Prakash, J.S.; Krishna, P.S.; Sirisha, K.; Kanesaki, Y.; Suzuki, I.; Shivaji, S. An RNA helicase, CrhR, regulates the low-temperature-inducible expression of heat-shock genes groES, groEL1 and groEL2 in Synechocystis sp. PCC 6803. Microbiology 2010, 156, 442–451. [Google Scholar]
- Rosana, A.R.; Ventakesh, M.; Chamot, D.; Patterson-Fortin, L.M.; Tarassova, O.; Espie, G.S. Inactivation of a low temperature-induced RNA helicase in Synechocystis sp. PCC 6803: Physiological and morphological consequences. 2012, 53, 646–658. [Google Scholar]
- Rowland, J.G.; Simon, W.J.; Prakash, J.S.; Slabas, A.R. Proteomics reveals a role for the RNA helicase crhR in the modulation of multiple metabolic pathways during cold acclimation of Synechocystis sp. PCC6803. J. Proteome. Res. 2011, 10, 3674–3689. [Google Scholar]
- Sireesha, K.; Radharani, B.; Krishna, P.S.; Sreedhar, N.; Subramanyam, R.; Mohanty, P. RNA helicase, CrhR is indispensable for the energy redistribution and the regulation of photosystem stoichiometry at low temperature in Synechocystis sp PCC6803. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 1525–1536. [Google Scholar]
- Tan, X.; Zhu, T.; Shen, S.; Yin, C.; Gao, H.; Xu, X. Role of Rbp1 in the acquired chill-light tolerance of cyanobacteria. J. Bacteriol. 2011, 193, 2675–2683. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar]
- Hartmut, K.L. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Methods in Enzymology; Academic Press: Waltham, MA, USA, 1987; pp. 350–382. [Google Scholar]
- Kondo, K.; Ochiai, Y.; Katayama, M.; Ikeuchi, M. The membrane-associated CpcG2-phycobilisome in Synechocystis: A new photosystem I antenna. Plant Physiol. 2007, 144, 1200–1210. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, H.; Chen, G.; Wang, Y.; Ding, Q.; Wei, D. Sll0528, a Site-2-Protease, Is Critically Involved in Cold, Salt and Hyperosmotic Stress Acclimation of Cyanobacterium Synechocystis sp. PCC 6803. Int. J. Mol. Sci. 2014, 15, 22678-22693. https://doi.org/10.3390/ijms151222678
Lei H, Chen G, Wang Y, Ding Q, Wei D. Sll0528, a Site-2-Protease, Is Critically Involved in Cold, Salt and Hyperosmotic Stress Acclimation of Cyanobacterium Synechocystis sp. PCC 6803. International Journal of Molecular Sciences. 2014; 15(12):22678-22693. https://doi.org/10.3390/ijms151222678
Chicago/Turabian StyleLei, Haijin, Gu Chen, Yuling Wang, Qinglong Ding, and Dong Wei. 2014. "Sll0528, a Site-2-Protease, Is Critically Involved in Cold, Salt and Hyperosmotic Stress Acclimation of Cyanobacterium Synechocystis sp. PCC 6803" International Journal of Molecular Sciences 15, no. 12: 22678-22693. https://doi.org/10.3390/ijms151222678