Three-Dimensional Structural Aspects of Protein–Polysaccharide Interactions
Abstract
:1. Introduction
2. Atomic Details of the Polylactosamine and Galectin Interaction
3. Structural Studies of CD44 and Hyaluronan (HA) Complex
4. Atomic Characterization of the Interaction between Carbohydrate-Binding Modules and β1–3 Glucan
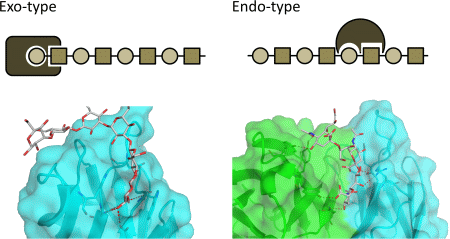
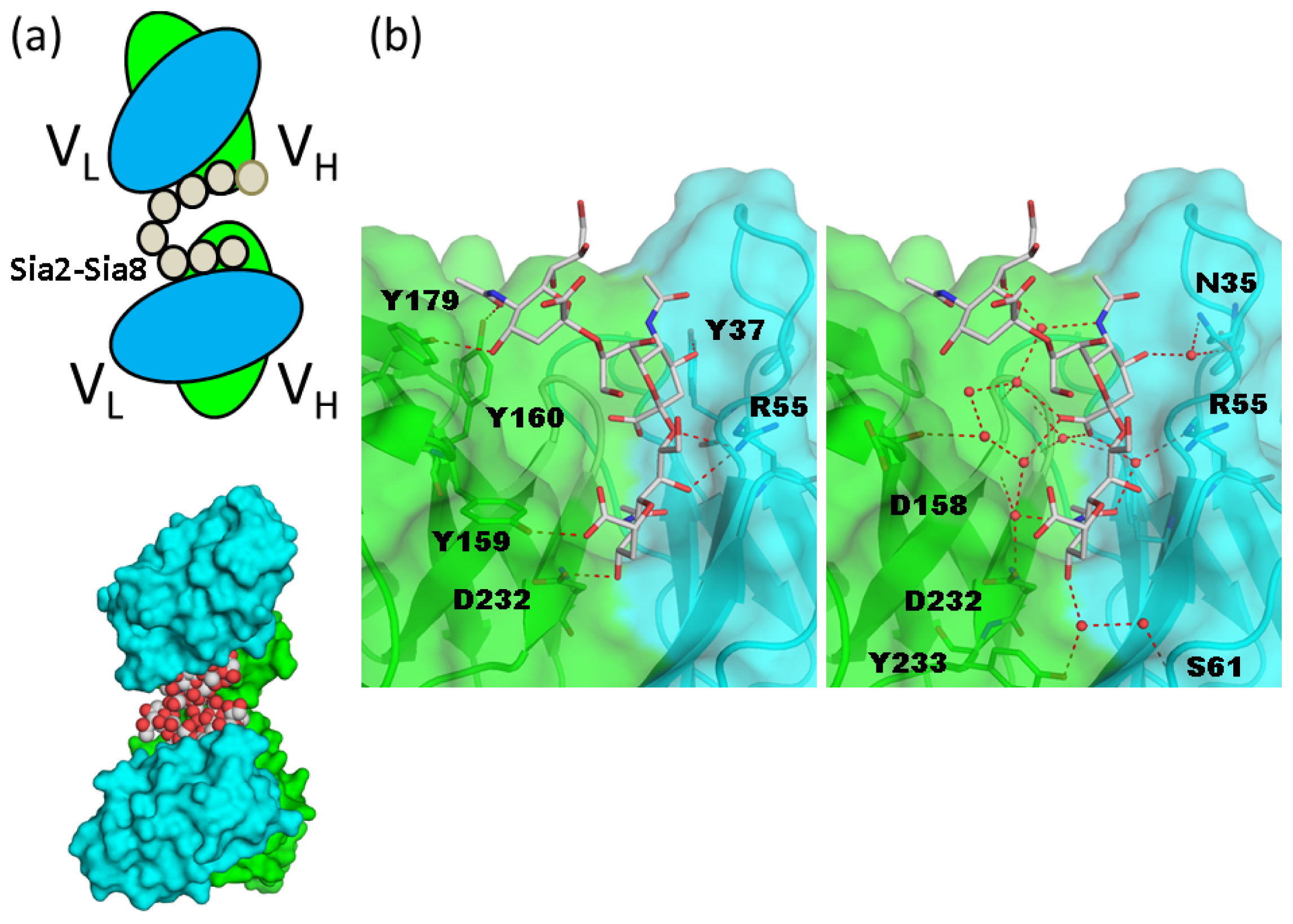
5. Interaction between Polysialic Acid and Antibodies
6. Additional Modifications
7. Conclusions and Perspectives
Acknowledgments
Conflicts of Interest
References
- Lee, R.T.; Lee, Y.C. Affinity enhancement by multivalent lectin-carbohydrate interaction. Glycoconj. J. 2000, 17, 543–551. [Google Scholar]
- Fred Brewer, C. Binding and cross-linking properties of galectins. Biochim. Biophys. Acta 2002, 1572, 255–262. [Google Scholar]
- Bourne, Y.; Bolgiano, B.; Liao, D.I.; Strecker, G.; Cantau, P.; Herzberg, O.; Feizi, T.; Cambillau, C. Crosslinking of mammalian lectin (galectin-1) by complex biantennary saccharides. Nat. Struct. Biol. 1994, 1, 863–870. [Google Scholar]
- Gupta, D.; Bhattacharyya, L.; Fant, J.; Macaluso, F.; Sabesan, S.; Brewer, C.F. Observation of unique cross-linked lattices between multiantennary carbohydrates and soybean lectin Presence of pseudo-2-fold axes of symmetry in complex type carbohydrates. Biochemistry 1994, 33, 7495– 7504. [Google Scholar]
- Olsen, L.R.; Dessen, A.; Gupta, D.; Sabesan, S.; Sacchettini, J.C.; Brewer, C.F. X-ray crystallographic studies of unique cross-linked lattices between four isomeric biantennary oligosaccharides and soybean agglutinin. Biochemistry 1997, 36, 15073–15080. [Google Scholar]
- Rabinovich, G.A.; Toscano, M.A.; Jackson, S.S.; Vasta, G.R. Functions of cell surface galectin–glycoprotein lattices. Curr. Opin. Struct. Biol. 2007, 17, 513–520. [Google Scholar]
- Georgelis, N.; Yennawar, N.H.; Cosgrove, D.J. Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin. Proc. Natl. Acad. Sci. USA 2012, 109, 14830–14835. [Google Scholar]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar]
- Boraston, A.B. The interaction of carbohydrate-binding modules with insoluble non-crystalline cellulose is enthalpically driven. Biochem. J. 2005, 385, 479–484. [Google Scholar]
- Houston, D.R.; Recklies, A.D.; Krupa, J.C.; van Aalten, D.M. Structure and ligand-induced conformational change of the 39-kDa glycoprotein from human articular chondrocytes. J. Biol. Chem. 2003, 278, 30206–30212. [Google Scholar]
- Fusetti, F.; Pijning, T.; Kalk, K.H.; Bos, E.; Dijkstra, B.W. Crystal structure and carbohydratebinding properties of the human cartilage glycoprotein-39. J. Biol. Chem. 2003, 278, 37753– 37760. [Google Scholar]
- Schimpl, M.; Rush, C.L.; Betou, M.; Eggleston, I.M.; Recklies, A.D.; van Aalten, D.M. Human YKL-39 is a pseudo-chitinase with retained chitooligosaccharide-binding properties. Biochem. J. 2012, 446, 149–157. [Google Scholar]
- Atrih, A.; Richardson, J.M.; Prescott, A.R.; Ferguson, M.A. Trypanosoma brucei glycoproteins contain novel giant poly-N-acetyllactosamine carbohydrate chains. J. Biol. Chem. 2005, 280, 865–871. [Google Scholar]
- Barondes, S.H.; Cooper, D.N.; Gitt, M.A.; Leffler, H. Galectins Structure and function of a large family of animal lectins. J. Biol. Chem. 1994, 269, 20807–20810. [Google Scholar]
- Hirabayashi, J.; Hashidate, T.; Arata, Y.; Nishi, N.; Nakamura, T.; Hirashima, M.; Urashima, T.; Oka, T.; Futai, M.; Muller, W.E.; et al. Oligosaccharide specificity of galectins: A search by frontal affinity chromatography. Biochim. Biophys. Acta 2002, 1572, 232–254. [Google Scholar]
- Nagae, M.; Nishi, N.; Murata, T.; Usui, T.; Nakamura, T.; Wakatsuki, S.; Kato, R. Structural analysis of the recognition mechanism of poly-N-acetyllactosamine by the human galectin-9 N-terminal carbohydrate recognition domain. Glycobiology 2009, 19, 112–117. [Google Scholar]
- Seetharaman, J.; Kanigsberg, A.; Slaaby, R.; Leffler, H.; Barondes, S.H.; Rini, J.M. X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 21-Å resolution. J. Biol. Chem. 1998, 273, 13047–13052. [Google Scholar]
- Leonidas, D.D.; Vatzaki, E.H.; Vorum, H.; Celis, J.E.; Madsen, P.; Acharya, K.R. Structural basis for the recognition of carbohydrates by human galectin-7. Biochemistry 1998, 37, 13930–13940. [Google Scholar]
- Kumar, S.; Frank, M.; Schwartz-Albiez, R. Understanding the specificity of human Galectin-8C domain interactions with its glycan ligands based on molecular dynamics simulations. PLoS One 2013, 8, e59761. [Google Scholar]
- Yoshida, H.; Yamashita, S.; Teraoka, M.; Itoh, A.; Nakakita, S.; Nishi, N.; Kamitori, S. X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains. FEBS J. 2012, 279, 3937–3951. [Google Scholar]
- Stowell, S.R.; Dias-Baruffi, M.; Penttilä, L.; Renkonen, O.; Nyame, A.K.; Cummings, R.D. Human galectin-1 recognition of poly-N-acetyllactosamine and chimeric polysaccharides. Glycobiology 2004, 14, 157–167. [Google Scholar]
- Di Virgilio, S.; Glushka, J.; Moremen, K.; Pierce, M. Enzymatic synthesis of natural and 13C enriched linear poly-N-acetyllactosamines as ligands for galectin-1. Glycobiology 1999, 9, 353– 364. [Google Scholar]
- Stannard, K.A.; Collins, P.M.; Ito, K.; Sullivan, E.M.; Scott, S.A.; Gabutero, E.; Darren Grice, I.; Low, P.; Nilsson, U.J.; Leffler, H.; et al. Galectin inhibitory disaccharides promote tumour immunity in a breast cancer model. Cancer Lett. 2010, 299, 95–110. [Google Scholar]
- Day, A.J.; Sheehan, J.K. Hyaluronan: Polysaccharide chaos to protein organisation. Curr. Opin. Struct. Biol. 2001, 11, 617–622. [Google Scholar]
- Day, A.J.; Prestwich, G.D. Hyaluronan-binding proteins: Tying up the giant. J. Biol. Chem. 2002, 277, 4585–4588. [Google Scholar]
- Day, A.J. The structure and regulation of hyaluronan-binding proteins. Biochem. Soc. Trans. 1999, 27, 115–121. [Google Scholar]
- Goodison, S.; Urquidi, V.; Tarin, D. CD44 cell adhesion molecules. Mol. Pathol. 1999, 52, 189– 196. [Google Scholar]
- Takeda, M.; Terasawa, H.; Sakakura, M.; Yamaguchi, Y.; Kajiwara, M.; Kawashima, H.; Miyasaka, M.; Shimada, I. Hyaluronan recognition mode of CD44 revealed by cross-saturation and chemical shift perturbation experiments. J. Biol. Chem. 2003, 278, 43550–43555. [Google Scholar]
- Teriete, P.; Banerji, S.; Noble, M.; Blundell, C.D.; Wright, A.J.; Pickford, A.R.; Lowe, E.; Mahoney, D.J.; Tammi, M.I.; Kahmann, J.D.; et al. Structure of the regulatory hyaluronan binding domain in the inflammatory leukocyte homing receptor CD44. Mol. Cell 2004, 13, 483–496. [Google Scholar]
- Takeda, M.; Ogino, S.; Umemoto, R.; Sakakura, M.; Kajiwara, M.; Sugahara, K.N.; Hayasaka, H.; Miyasaka, M.; Terasawa, H.; Shimada, I. Ligand-induced structural changes of the CD44 hyaluronan-binding domain revealed by NMR. J. Biol. Chem. 2006, 281, 40089–40095. [Google Scholar]
- Banerji, S.; Wright, A.J.; Noble, M.; Mahoney, D.J.; Campbell, I.D.; Day, A.J.; Jackson, D.G. Structures of the CD44–hyaluronan complex provide insight into a fundamental carbohydrate–protein interaction. Nat. Struct. Mol. Biol. 2007, 14, 234–239. [Google Scholar]
- Ogino, S.; Nishida, N.; Umemoto, R.; Suzuki, M.; Takeda, M.; Terasawa, H.; Kitayama, J.; Matsumoto, M.; Hayasaka, H.; Miyasaka, M.; et al. Two-state conformations in the hyaluronan-binding domain regulate CD44 adhesiveness under flow condition. Structure 2010, 18, 649–656. [Google Scholar]
- Lesley, J.; Hascall, V.C.; Tammi, M.; Hyman, R. Hyaluronan binding by cell surface CD44. J. Biol. Chem. 2000, 275, 26967–26975. [Google Scholar]
- Lesley, J.; Gál, I.; Mahoney, D.J.; Cordell, M.R.; Rugg, M.S.; Hyman, R.; Day, A.J.; Mikecz, K. TSG-6 modulates the interaction between hyaluronan and cell surface CD44. J. Biol. Chem. 2004, 279, 25745–25754. [Google Scholar]
- Blundell, C.D.; Mahoney, D.J.; Almond, A.; DeAngelis, P.L.; Kahmann, J.D.; Teriete, P.; Pickford, A.R.; Campbell, I.D.; Day, A.J. The link module from ovulation- and inflammationassociated protein TSG-6 changes conformation on hyaluronan binding. J. Biol. Chem. 2003, 278, 49261–49270. [Google Scholar]
- Chuah, C.T.; Sarko, A.; Deslandes, Y.; Marchessault, R.H. Packing analysis of carbohydrates and polysaccharides14 Triple-helical crystalline-structure of curdlan and paramylon hydrates. Macromolecules 1983, 16, 1375–1382. [Google Scholar]
- Sletmoen, M.; Stokke, B.T. Higher order structure of (13)-b-d-glucans and its influence on their biological activities and complexation abilities. Biopolymers 2008, 89, 310–321. [Google Scholar]
- Saito, H.; Yoshioka, Y.; Uehara, N.; Aketagawa, J.; Tanaka, S.; Shibata, Y. Relationship between conformation and biological response for (1→3)-β-d-glucans in the activation of coagulation factor G from limulus amebocyte lysate and host-mediated antitumor activity Demonstration of single-helix conformation as a stimulant. Carbohydr. Res. 1991, 217, 181–190. [Google Scholar]
- Boraston, A.B.; Nurizzo, D.; Notenboom, V.; Ducros, V.; Rose, D.R.; Kilburn, D.G.; Davies, G.J. Differential oligosaccharide recognition by evolutionarily-related β-14 and β-13 glucan-binding modules. J. Mol. Biol. 2002, 319, 1143–1156. [Google Scholar]
- Van Bueren, A.L.; Morland, C.; Gilbert, H.J.; Boraston, A.B. Family 6 carbohydrate binding modules recognize the non-reducing end of β-13-linked glucans by presenting a unique ligand binding surface. J. Biol. Chem. 2005, 280, 530–537. [Google Scholar]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar]
- Takahasi, K.; Ochiai, M.; Horiuchi, M.; Kumeta, H.; Ogura, K.; Ashida, M.; Inagaki, F. Solution structure of the silkworm βGRP/GNBP3 N-terminal domain reveals the mechanism for β-13-glucan-specific recognition. Proc. Natl. Acad. Sci. USA 2009, 106, 11679–11684. [Google Scholar]
- Kanagawa, M.; Satoh, T.; Ikeda, A.; Adachi, Y.; Ohno, N.; Yamaguchi, Y. Structural insights into recognition of triple-helical β-glucans by an insect fungal receptor. J. Biol. Chem. 2011, 286, 29158–29165. [Google Scholar]
- Dai, H.; Hiromasa, Y.; Takahashi, D.; VanderVelde, D.; Fabrick, J.A.; Kanost, M.R.; Krishnamoorthi, R. An initial event in the insect innate immune response: Structural and biological studies of interactions between β-13-glucan and the N-terminal domain of β-13-glucan recognition protein. Biochemistry 2013, 52, 161–170. [Google Scholar]
- Palma, A.S.; Feizi, T.; Zhang, Y.; Stoll, M.S.; Lawson, A.M.; Díaz-Rodríguez, E.; Campanero-Rhodes, M.A.; Costa, J.; Gordon, S.; Brown, G.D.; et al. Ligands for the β-glucan receptor Dectin-1 assigned using “designer” microarrays of oligosaccharide probes (neoglycolipids) generated from glucan polysaccharides. J. Biol. Chem. 2006, 281, 5771–5779. [Google Scholar]
- Hanashima, S.; Ikeda, A.; Tanaka, H.; Adachi, Y.; Ohno, N.; Takahashi, T.; Yamaguchi, Y. NMR study of short β(1–3)-glucans provides insights into the structure and interaction with Dectin-1. Glycoconj. J. 2013. [Google Scholar] [CrossRef]
- Meagher, J.L.; Winter, H.C.; Ezell, P.; Goldstein, I.J.; Stuckey, J.A. Crystal structure of banana lectin reveals a novel second sugar binding site. Glycobiology 2005, 15, 1033–1042. [Google Scholar]
- Nakata, D.; Troy, F.A., 2nd. Degree of polymerization (DP) of polysialic acid (polySia) on neural cell adhesion molecules (N-CAMS): Development and application of a new strategy to accurately determine the DP of polySia chains on N-CAMS. J. Biol. Chem. 2005, 280, 38305–38316. [Google Scholar]
- Sato, C.; Kitajima, K. Disialic oligosialic and polysialic acids: Distribution functions and related disease. J. Biochem. 2013, 154, 115–136. [Google Scholar]
- Finne, J. Occurrence of unique polysialosyl carbohydrate units in glycoproteins of developing brain. J. Biol. Chem. 1982, 257, 11966–11970. [Google Scholar]
- Hanashima, S.; Sato, C.; Tanaka, H.; Takahashi, T.; Kitajima, K.; Yamaguchi, Y. NMR study into the mechanism of recognition of the degree of polymerization by oligo/polysialic acid antibodies. Bioorg. Med. Chem. 2013, 21, 6069–6076. [Google Scholar]
- Nagae, M.; Ikeda, A.; Hane, M.; Hanashima, S.; Kitajima, K.; Sato, C.; Yamaguchi, Y. Crystal structure of anti-polysialic acid antibody single chain Fv fragment complexed with octasialic acid: Insight into the binding preference for polysialic acid. J. Biol. Chem. 2013, 288, 33784–33796. [Google Scholar]
- Evans, S.V.; Sigurskjold, B.W.; Jennings, H.J.; Brisson, J.R.; To, R.; Tse, W.C.; Altman, E.; Frosch, M.; Weisgerber, C.; Kratzin, H.D.; et al. Evidence for the extended helical nature of polysaccharide epitopes The 28 Å resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for α-(2→8)-polysialic acid. Biochemistry 1995, 34, 6737–6744. [Google Scholar]
- Yamasaki, R.; Bacon, B. Three-dimensional structural analysis of the group B polysaccharide of Neisseria meningitidis 6275 by two-dimensional NMR: The polysaccharide is suggested to exist in helical conformations in solution. Biochemistry 1991, 30, 851–857. [Google Scholar]
- Brisson, J.R.; Baumann, H.; Imberty, A.; Pérez, S.; Jennings, H.J. Helical epitope of the group B meningococcal α(2–8)-linked sialic acid polysaccharide. Biochemistry 1992, 31, 4996–5004. [Google Scholar]
- Yongye, A.B.; Gonzalez-Outeiriño, J.; Glushka, J.; Schultheis, V.; Woods, R.J. The conformational properties of methyl α-(28)-di/trisialosides and their N-acyl analogues: Implications for anti-Neisseria meningitidis B vaccine design. Biochemistry 2008, 47, 12493–12514. [Google Scholar]
- Henderson, T.J.; Venable, R.M.; Egan, W. Conformational flexibility of the group B meningococcal polysaccharide in solution. J. Am. Chem. Soc. 2003, 125, 2930–2939. [Google Scholar]
- Jin, L.; Abrahams, J.P.; Skinner, R.; Petitou, M.; Pike, R.N.; Carrell, R.W. The anticoagulant activation of antithrombin by heparin. Proc. Natl. Acad. Sci. USA 1997, 94, 14683–14688. [Google Scholar]
- Faham, S.; Hileman, R.E.; Fromm, J.R.; Linhardt, R.J.; Rees, D.C. Heparin structure and interactions with basic fibroblast growth factor. Science 1996, 271, 1116–1120. [Google Scholar]
- Capila, I.; Hernáiz, M.J.; Mo, Y.D.; Mealy, T.R.; Campos, B.; Dedman, J.R.; Linhardt, R.J.; Seaton, B.A. Annexin V–heparin oligosaccharide complex suggests heparan sulfate-mediated assembly on cell surfaces. Structure 2001, 9, 57–64. [Google Scholar]
- Ono, S.; Hane, M.; Kitajima, K.; Sato, C. Novel regulation of fibroblast growth factor 2 (FGF2)-mediated cell growth by polysialic acid. J. Biol. Chem. 2012, 287, 3710–3722. [Google Scholar]





© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nagae, M.; Yamaguchi, Y. Three-Dimensional Structural Aspects of Protein–Polysaccharide Interactions. Int. J. Mol. Sci. 2014, 15, 3768-3783. https://doi.org/10.3390/ijms15033768
Nagae M, Yamaguchi Y. Three-Dimensional Structural Aspects of Protein–Polysaccharide Interactions. International Journal of Molecular Sciences. 2014; 15(3):3768-3783. https://doi.org/10.3390/ijms15033768
Chicago/Turabian StyleNagae, Masamichi, and Yoshiki Yamaguchi. 2014. "Three-Dimensional Structural Aspects of Protein–Polysaccharide Interactions" International Journal of Molecular Sciences 15, no. 3: 3768-3783. https://doi.org/10.3390/ijms15033768