Access of Hydrogen-Radicals to the Peptide-Backbone as a Measure for Estimating the Flexibility of Proteins Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry
Abstract
:1. Introduction
2. Results and Discussion
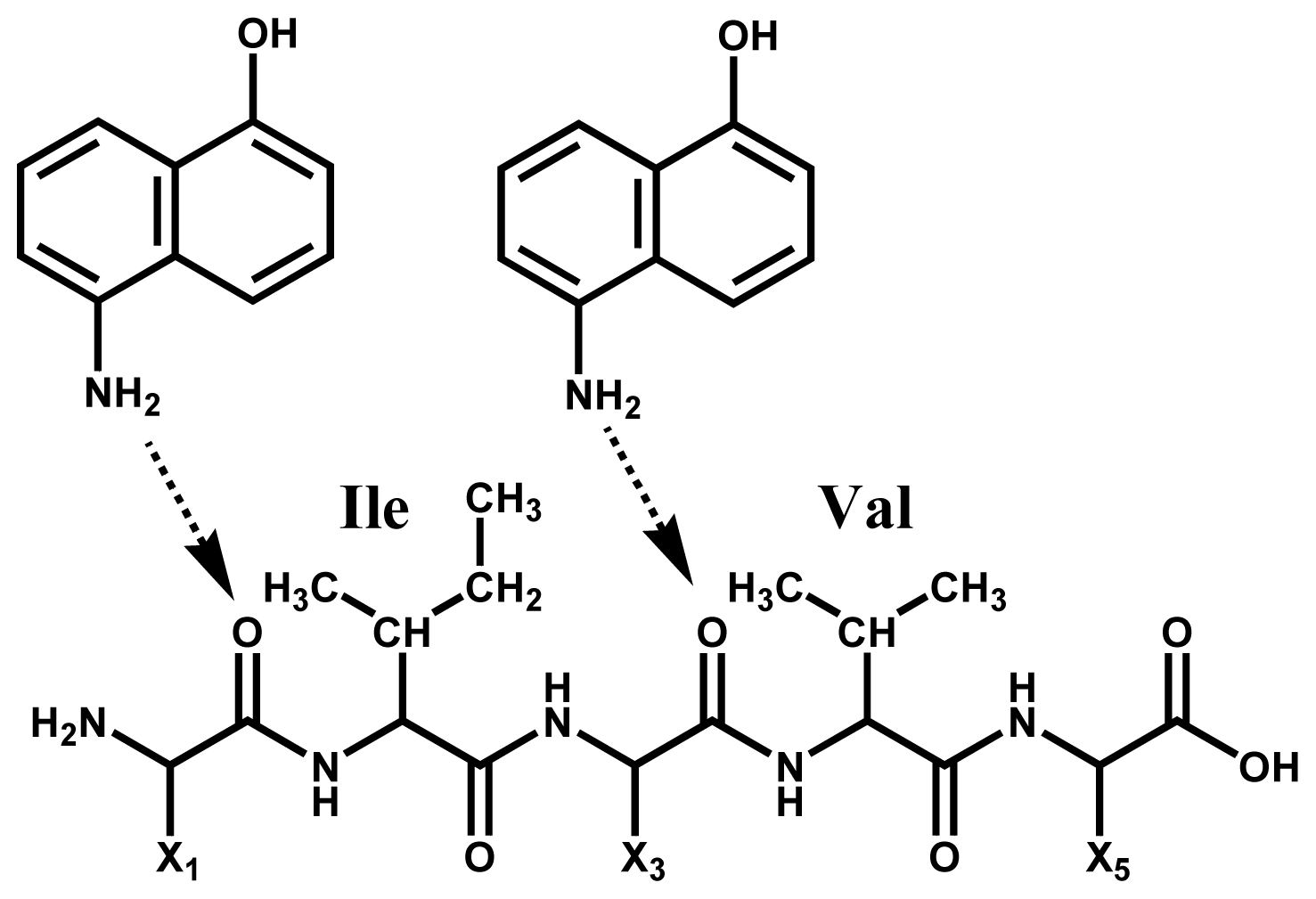
2.1. Preferential Binding of Hydrogen-Radicals Determining One-Side Preferential Cleavage at N-Cα Bond of Peptide Backbone
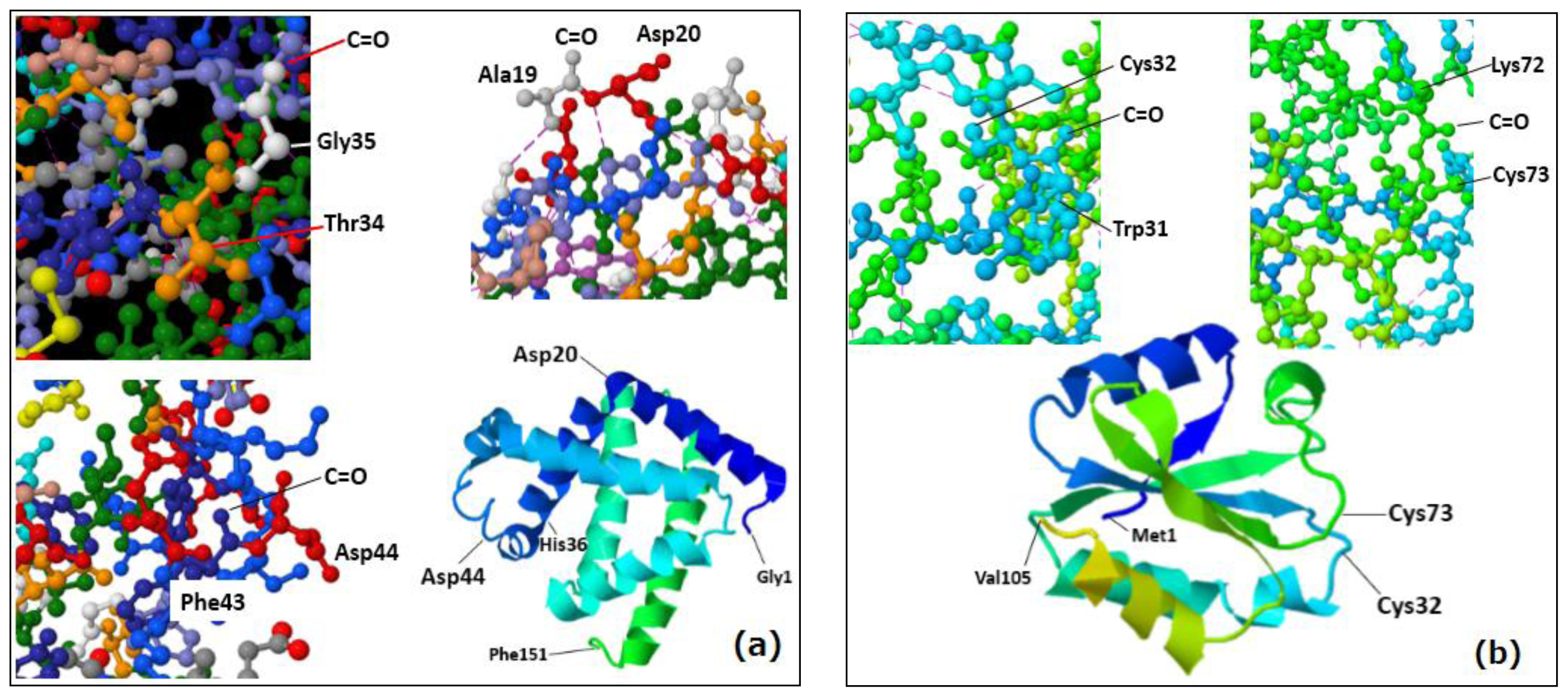
2.2. Discontinuous Intense ISD (In-Source Decay) Ion Peaks Reflect Helix-Free Regions of Protein
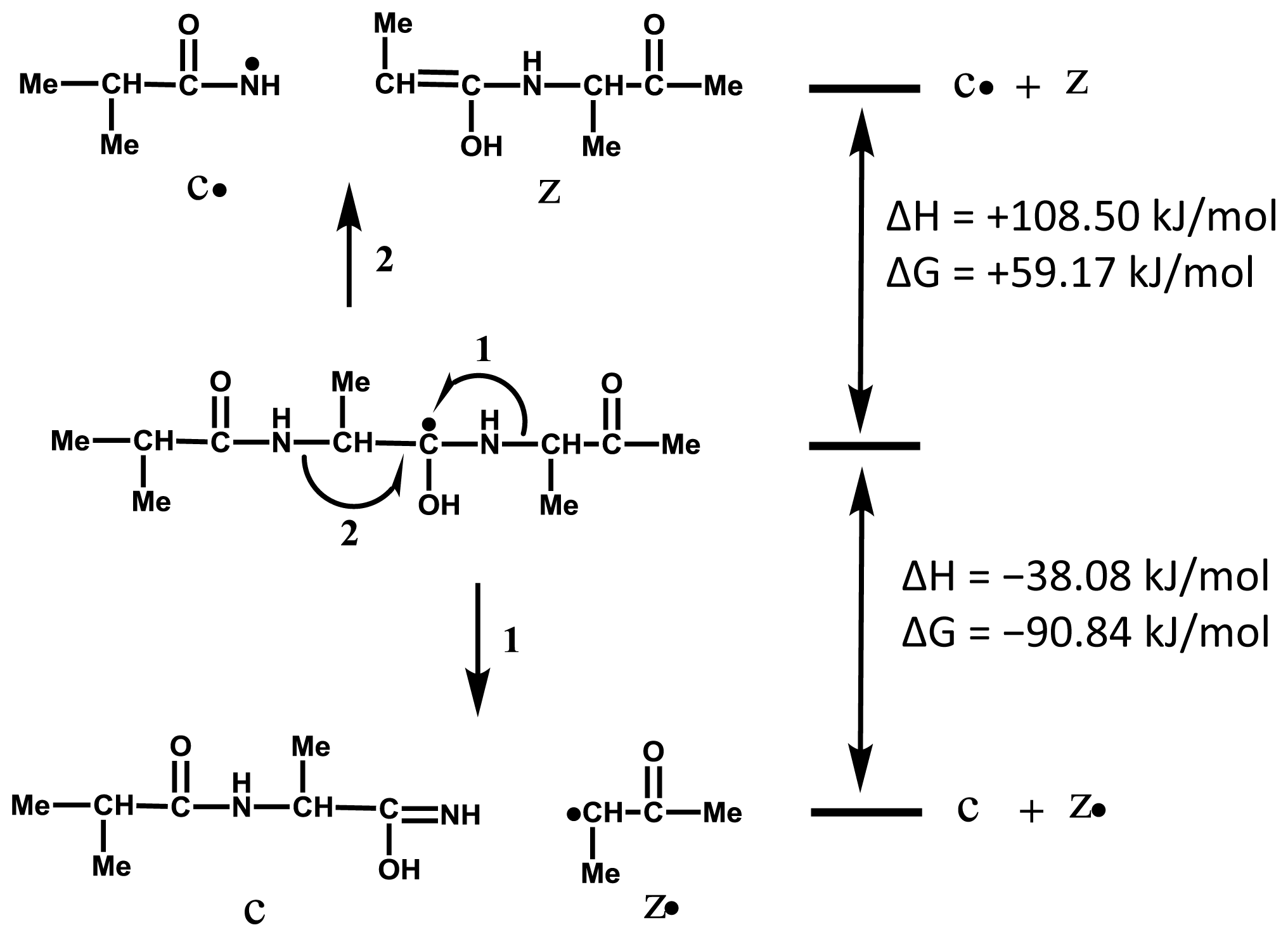
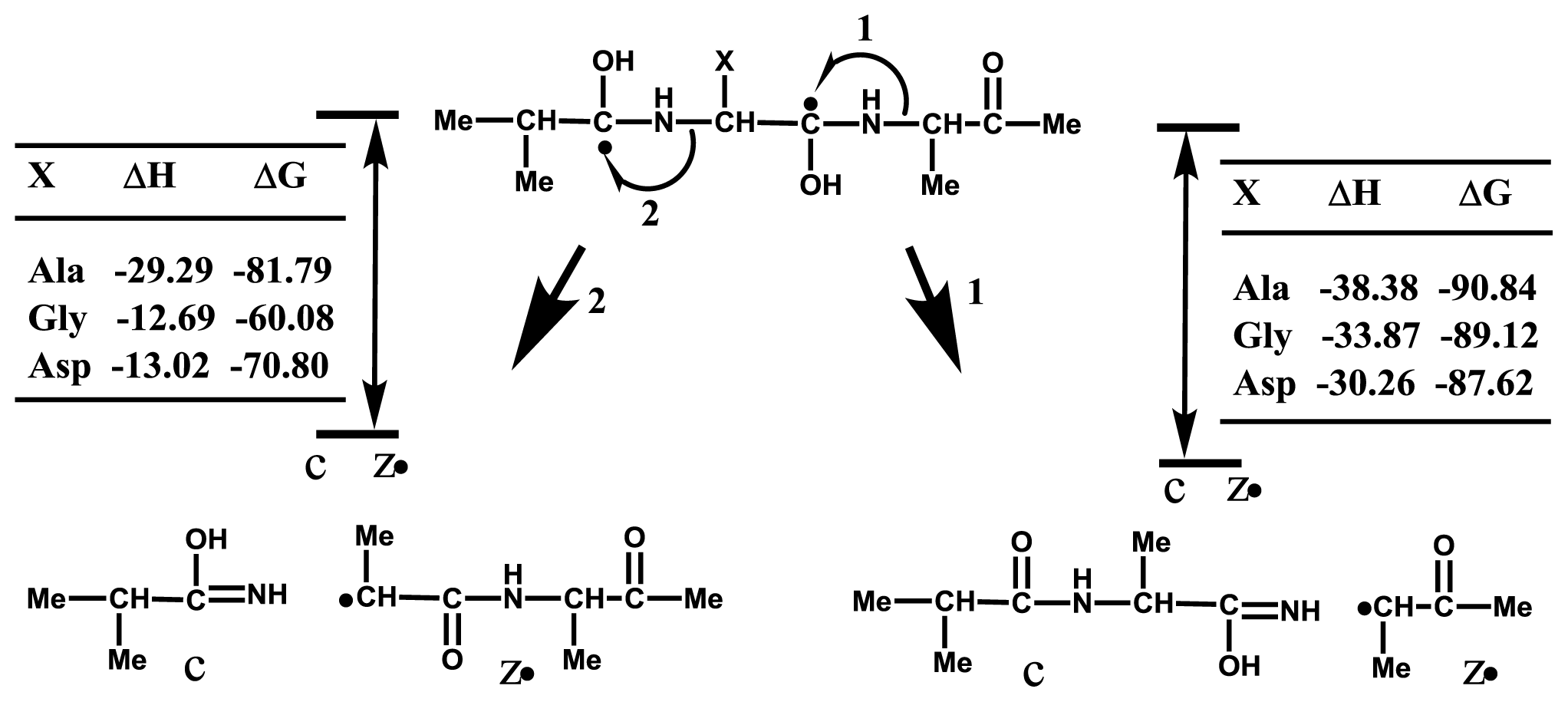
2.3. Accessibility of Carbonyl Oxygens of the Backbone to Hydrogen-Radicals
2.4. Discussion
3. Materials and Methods
3.1. Chemicals
3.2. Mass Spectrometry
3.3. Sample Preparation
3.4. Ab Initio Calculations
4. Conclusions
Supplementary Information
ijms-15-08428-s001.pdfAcknowledgments
Conflicts of Interest
- Author ContributionsM.T.—Designed and interpreted all experimental data, prepared the final manuscript, K.N.—Performed MS experiments and analyzed MS spectra, R.I.—Performed ab initio calculations, K.I.—Performed MS experiments. All authors read and approved the final manuscript.
References
- Karas, M.; Bachmann, D.; Bahr, U.; Hillenkamp, F. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int. J. Mass Spectrom. Ion. Proc 1987, 78, 53–68. [Google Scholar]
- Tanaka, K.; Waki, H.; Ido, Y.; Akita, S.; Yoshida, Y.; Yoshida, T. Protein and polymer analyses up to m/z 100,000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom 1988, 2, 151–153. [Google Scholar]
- Whitehouse, C.M.; Dreyer, R.N.; Yamashita, M.; Fenn, J.B. Electrospray interface for liquid chromatographs and mass spectrometers. Anal. Chem 1985, 57, 675–679. [Google Scholar]
- Fenn, J.B.; Mann, M.; Meng, C.K.; Wong, S.F.; Whitehouse, C.M. Electrospray ionization for mass spectrometry of large biomolecules. Science 1989, 246, 64–71. [Google Scholar]
- Rand, K.D.; Bache, N.; Nedertoft, M.M.; Jorgensen, J.D. Spatially resolved protein hydrogen exchange measured by matrix-assisted laser desorption ionization in-source decay. Anal. Chem 2011, 83, 8859–8862. [Google Scholar]
- Takayama, M. Flexible Xxx-Asp/Asn and Gly-Xxx residues of equine cytochrome c in matrix-assisted laser desorption/ionization in-source decay mass spectrometry. Mass Spectrom 2012. [Google Scholar] [CrossRef]
- Sterling, H.J.; Williams, E.R. Real-time hydrogen/deuterium exchange kinetics via supercharged electrospray ionization tandem mass spectrometry. Anal. Chem 2010, 82, 9050–9057. [Google Scholar]
- Uversky, V.V.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar]
- Karplus, P.A.; Schulz, G.E. Prediction of chain flexibility in proteins. Naturwissenschaften 1985, 72, 212–213. [Google Scholar]
- Radivojac, P.; Obradoic, Z.; Smith, D.K.; Zhu, G.; Vucetic, S.; Brown, C.J.; Lawson, J.D.; Dunker, A.K. Protein flexibility and intrinsic disorder. Protein Sci 2004, 13, 71–80. [Google Scholar]
- Chow, P.Y.; Fasman, G.D. Prediction of β-turns. Biophys. J 1979, 26, 367–384. [Google Scholar]
- Williams, R.W.; Chang, A.; Juretic, D.; Loughran, S. Secondary structure predictions and medium range interactions. Biochim. Biophys. Acta 1987, 916, 200–204. [Google Scholar]
- Bougault, C.; Feng, L.; Glushka, J.; Kupce, E.; Prestegard, J.H. Quantitation of rapid proton-deuteron amide exchange using hadamsrd spectroscopy. J. Biomol. NMR 2004, 28, 385–390. [Google Scholar]
- Huang, F.; Nau, W.M. A conformational flexibility scale for amino acids in proteins. Angew. Chem. Int. Ed 2003, 42, 2269–2272. [Google Scholar]
- Smith, D.K.; Radivojac, P.; Obradovic, Z.; Dunker, A.K.; Zhu, G. Improved amino acid flexibility parameters. Protein Sci 2003, 12, 1060–1107. [Google Scholar]
- Milne, J.S.; Mayne, L.; Roder, H.; Wand, A.J.; Englander, S.W. Determinations of protein hydrogen exchange studied in equine cytochrome c. Protein Sci 1998, 7, 739–745. [Google Scholar]
- Iimuro, R.; Takayama, M. Analysis of flexibility of proteins by means of positive and negative ion MALDI in-Source decay mass spectrometry. Mass Spectrom 2014. [Google Scholar] [CrossRef]
- Takayama, M.; Sekiya, S.; Iimuro, R.; Iwamoto, S.; Tanaka, K. Selective and nonselective cleavages in positive and negative CID of the fragments generated from in-source decay of intact proteins in MALDI-MS. J. Am. Soc. Mass Spectrom 2014, 25, 120–131. [Google Scholar]
- Demeure, K.; Quinton, L.; Gabelica, V.; de Pauw, E. Rational selection of the optimum MALDI matrix for top-down proteomics by in-source decay. Anal. Chem 2008, 79, 8678–8685. [Google Scholar]
- Takayama, M. N-Cα bond cleavage of the peptide backbone via hydrogen abstraction. J. Am. Soc. Mass Spectrom 2001, 12, 1044–1049. [Google Scholar]
- Kocher, T.; Engstrom, A.; Zubarev, R.A. Fragmentation of peptides in MALDI in-source decay mediated by hydrogen radicals. Anal. Chem 2005, 77, 172–177. [Google Scholar]
- Hardouin, J. Protein sequence information by matrix-assisted laser desorption/ionization in-source decay mass spectrometry. Mass Spectrom. Rev 2007, 26, 672–682. [Google Scholar]
- Takayama, M.; Osaka, I.; Sakakura, M. Influence of secondary structure on in-source decay of protein inn matrix-assisted laser desorption/ionization mass spectrometry. Mass Spectrom 2012. doi: 105702/massspectrometry.A0001. [Google Scholar]
- Osaka, I.; Sakai, M.; Takayama, M. 5-Amino-1-naphthol, a novel 1,5-naphthalene derivative matrix suitable for matrix-assisted laser desorption/ionization in-source decay of phosphorylated peptides. Rapid Commun. Mass Spectrom 2013, 27, 103–108. [Google Scholar]
- Takayama, M.; Tsugita, A. Does in-source decay occur independent of the ionization process in matrix-assisted laser desorption? Int. J. Mass Spectrom 1998, 181, L1–L6. [Google Scholar]
- Demeure, K.; Gabelica, V.; de Pauw, E. New advances in the understanding of the in-source decay in MALDI-TOF-MS. J. Am. Soc. Mass Spectrom 2010, 21, 1906–1191. [Google Scholar]
- Nishikaze, T.; Takayama, M. Study of factors governing negative molecular ion yields of amino acid and peptide in FAB, MALDI and ESI mass spectrometry. Int. J. Mass Spectrom 2007, 268, 47–59. [Google Scholar]
- Creiton, T.E. Mechanisms of Protein Folding; Pain, R.H., Ed.; IRL Press: Oxford, UK, 1994; pp. 1–25. [Google Scholar]
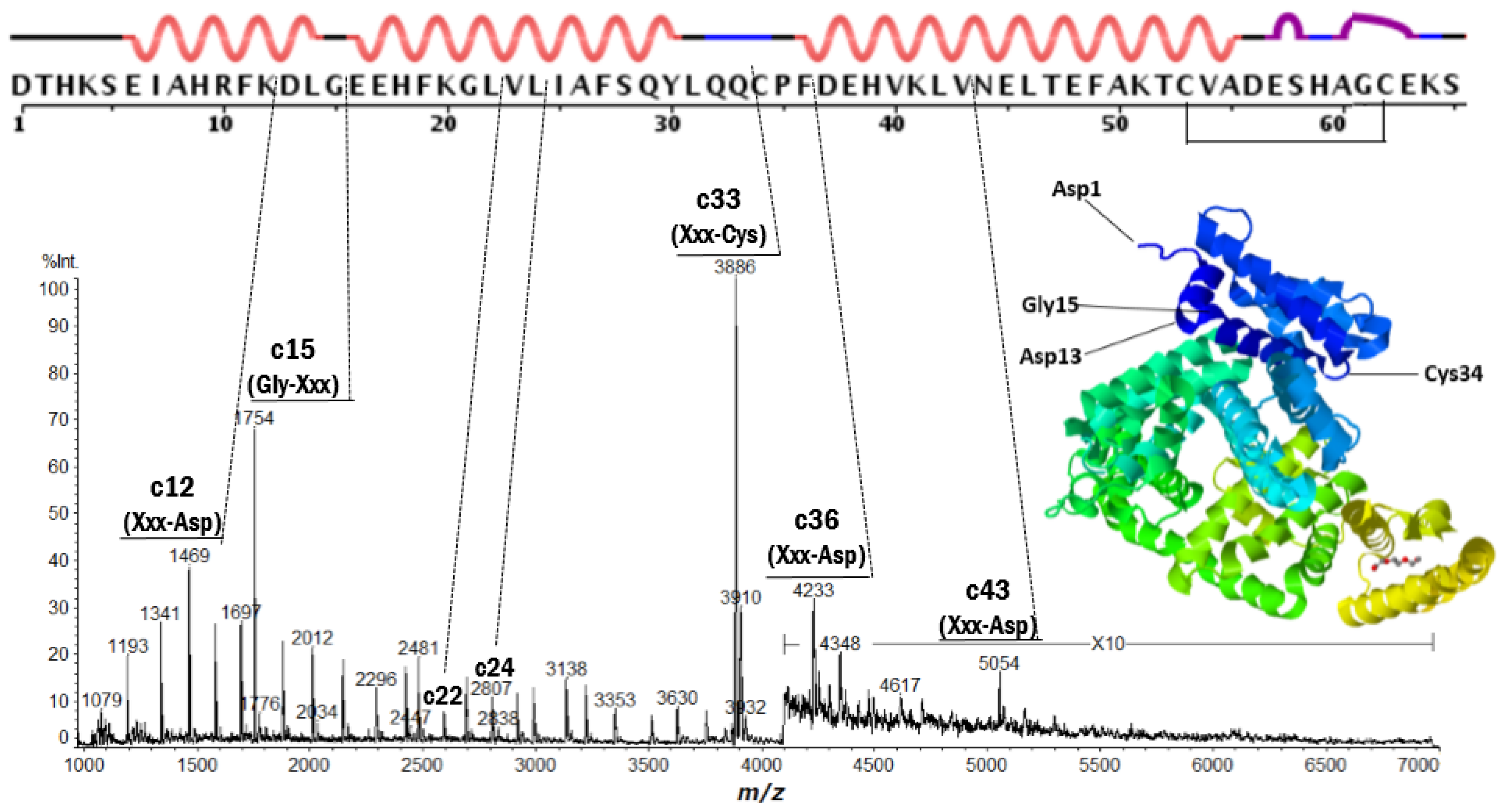
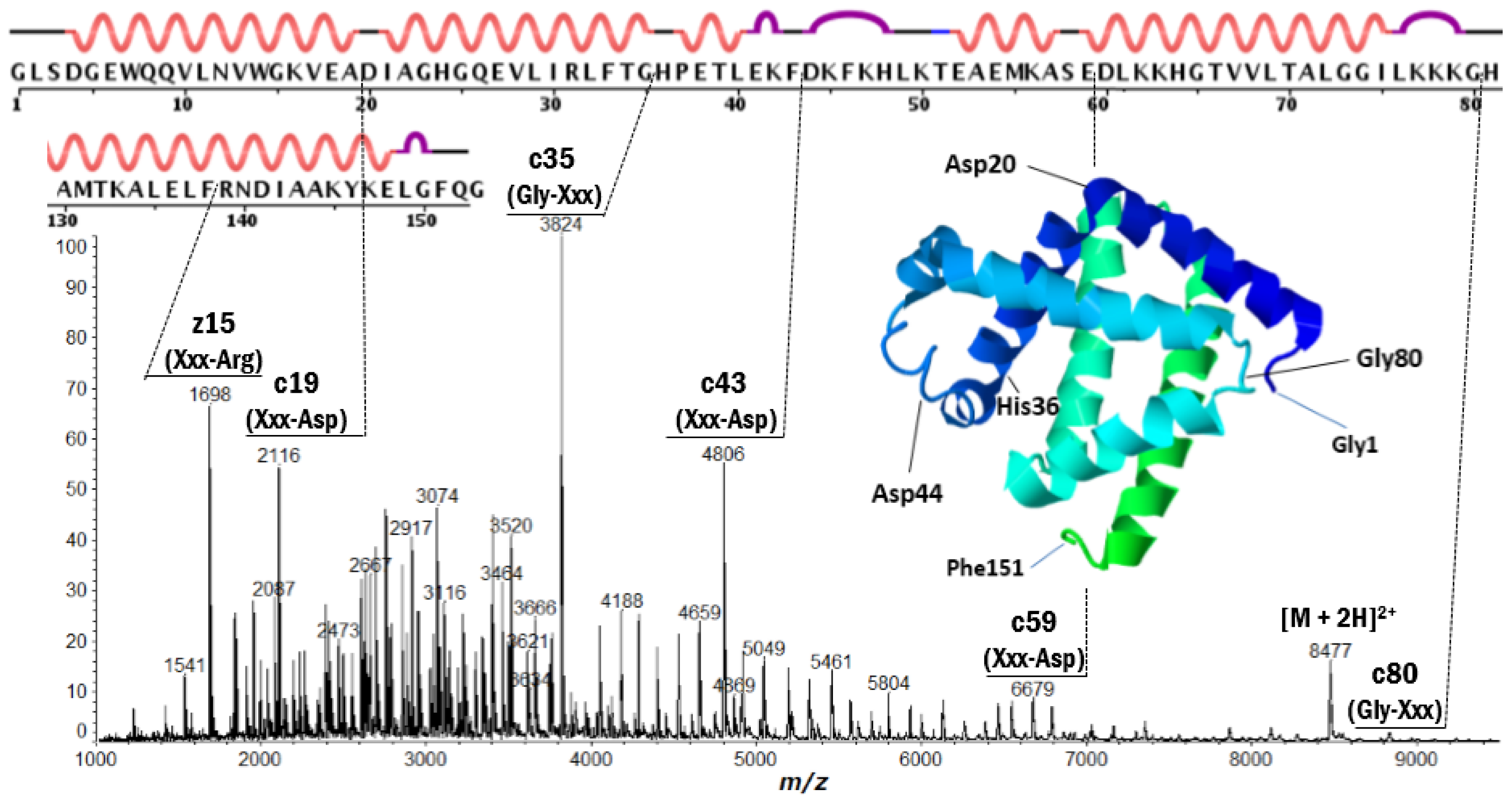
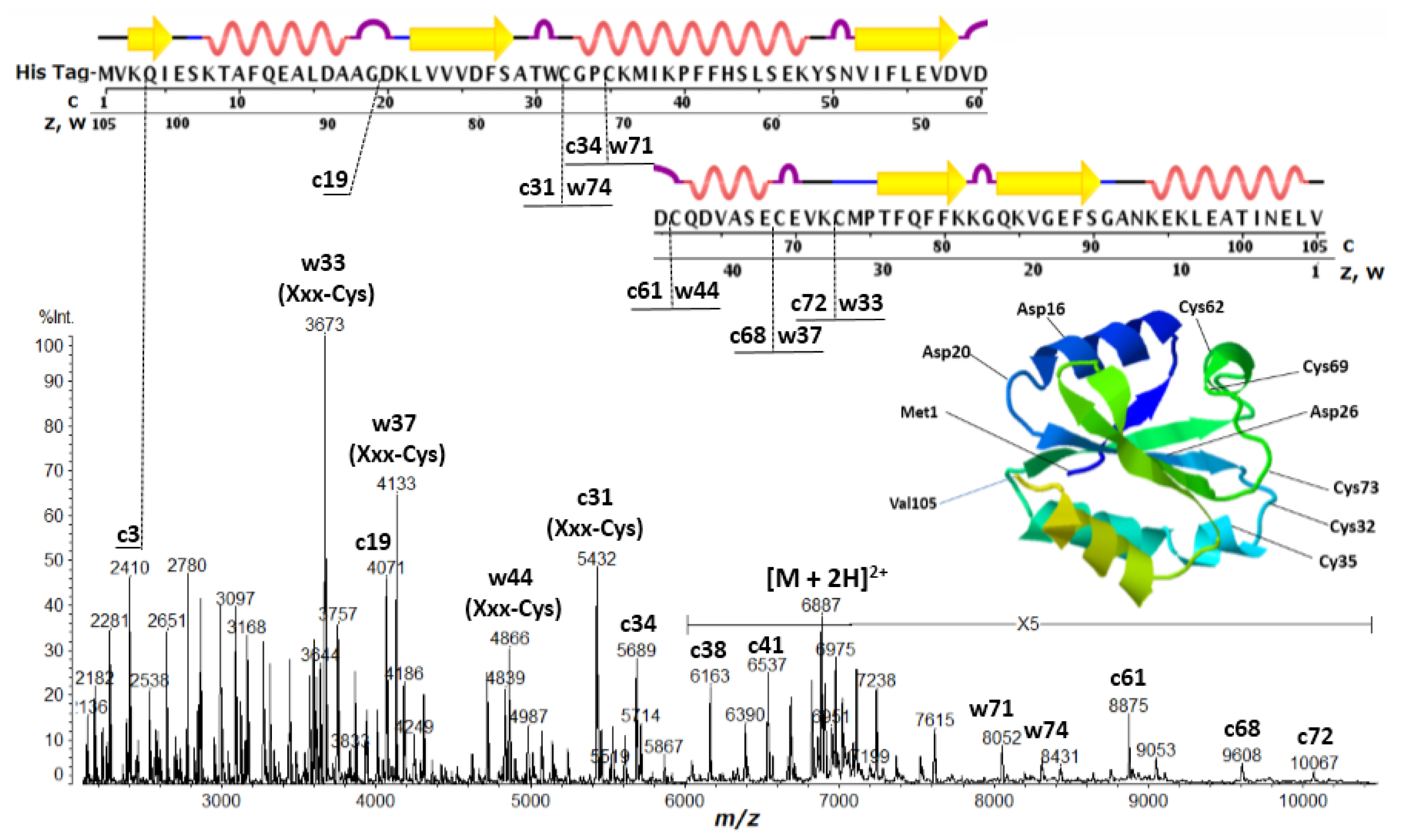

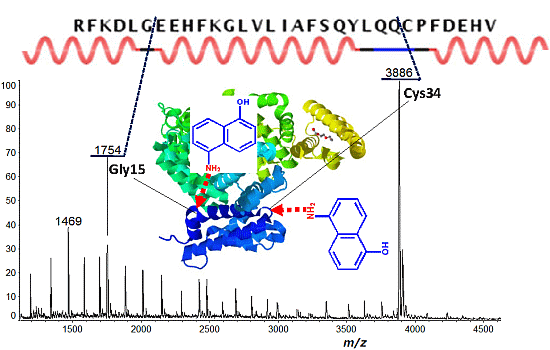
| Protein | Fragment Ion and Cleavage Residues |
|---|---|
| BSA | c12 (Xxx-Asp), c15 (Gly-Xxx), c33 (Xxx-Cys) |
| Myoglobin | z′15 (Xxx-Arg), c19 (Xxx-Asp), c35 (Gly-Xxx), c43 (Xxx-Asp), c59 (Xxx-Asp), c80 (Gly-Xxx) |
| Thioredoxin | c3 (Lys-Xxx), c19 (Gly-Xxx), c25 (Xxx-Asp), c31 (Xxx-Cys), c34 (Xxx-Cys), c61 (Xxx-Cys), c68 (Xxx-Cys), c72 (Xxx-Cys), w33 (Xxx-Cys), w37 (Xxx-Cys), w44 (Xxx-Cys), w71 (Xxx-Cys), w74 (Xxx-Cys) |
| Factor for Protein Flexibility | Flexible Amino Acid Residue |
|---|---|
| B-factor [10] | Asp, Asn, Gly, Pro, Lys, Glu, Gln, Ser |
| Turn preference [11] | Asp, Asn, Gly, Cys, Pro, Ser |
| Protection [13] | Asp, Asn, Gly, Lys, Thr, Ile, Met |
| Fluorescence decay [14] | Asp, Asn, Gly, Ser, Ala |
| Hydrogen-radical accessibility | Asp, Asn, Gly, Cys |
© 2014 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Takayama, M.; Nagoshi, K.; Iimuro, R.; Inatomi, K. Access of Hydrogen-Radicals to the Peptide-Backbone as a Measure for Estimating the Flexibility of Proteins Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Int. J. Mol. Sci. 2014, 15, 8428-8442. https://doi.org/10.3390/ijms15058428
Takayama M, Nagoshi K, Iimuro R, Inatomi K. Access of Hydrogen-Radicals to the Peptide-Backbone as a Measure for Estimating the Flexibility of Proteins Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. International Journal of Molecular Sciences. 2014; 15(5):8428-8442. https://doi.org/10.3390/ijms15058428
Chicago/Turabian StyleTakayama, Mitsuo, Keishiro Nagoshi, Ryunosuke Iimuro, and Kazuma Inatomi. 2014. "Access of Hydrogen-Radicals to the Peptide-Backbone as a Measure for Estimating the Flexibility of Proteins Using Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry" International Journal of Molecular Sciences 15, no. 5: 8428-8442. https://doi.org/10.3390/ijms15058428