SUMOylation of FOXM1B Alters Its Transcriptional Activity on Regulation of MiR-200 Family and JNK1 in MCF7 Human Breast Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. Forkhead Box Protein M1 B (FOXM1B) Is a Substrate for Modification by Small Ubiquitin-Related Modifier (SUMO)
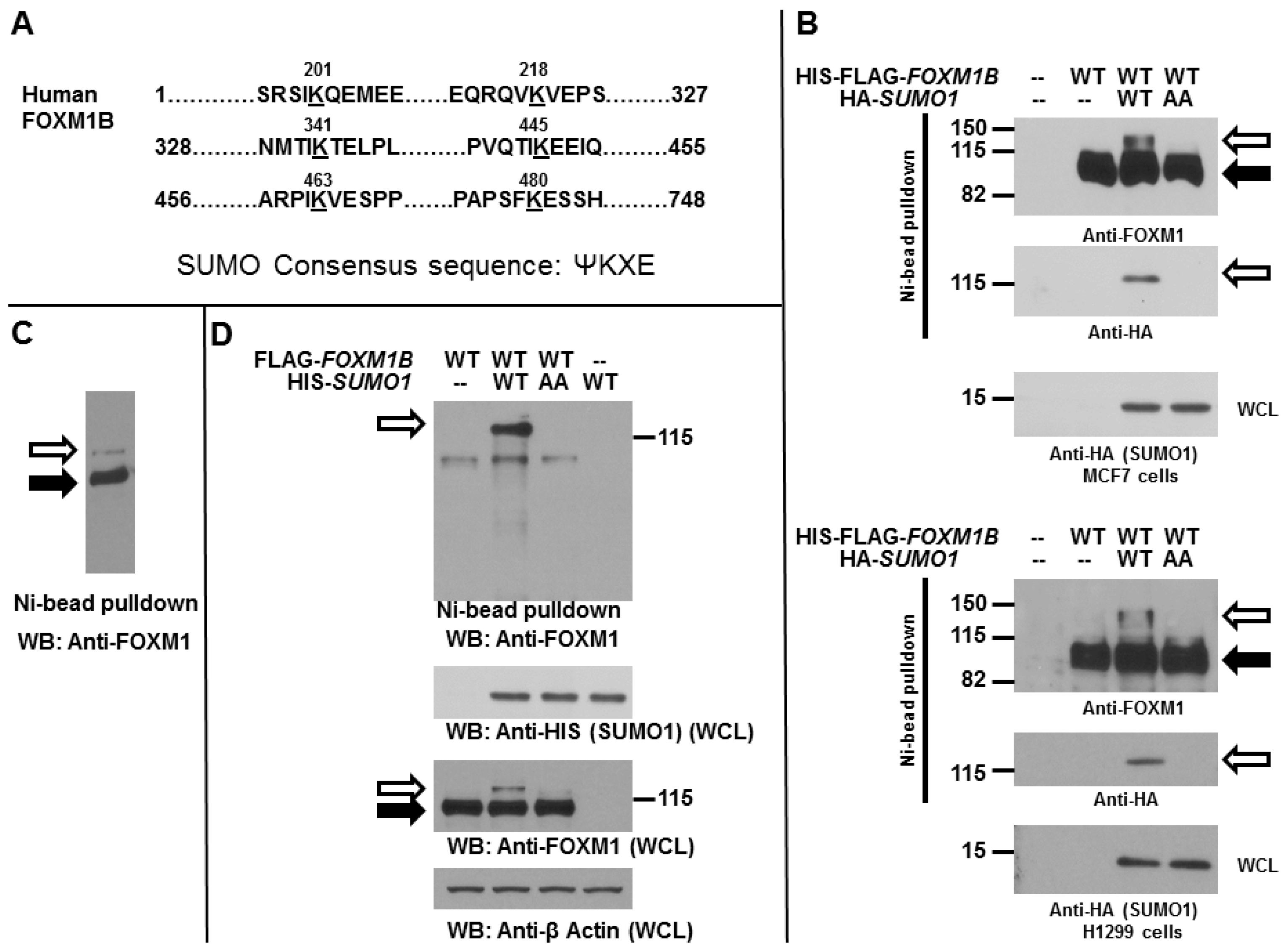
2.2. SUMOylation of FOXM1B Is Modulated by SENP2 and PIASy

2.3. Lysine 463 Is the Major SUMO Site in FOXM1B
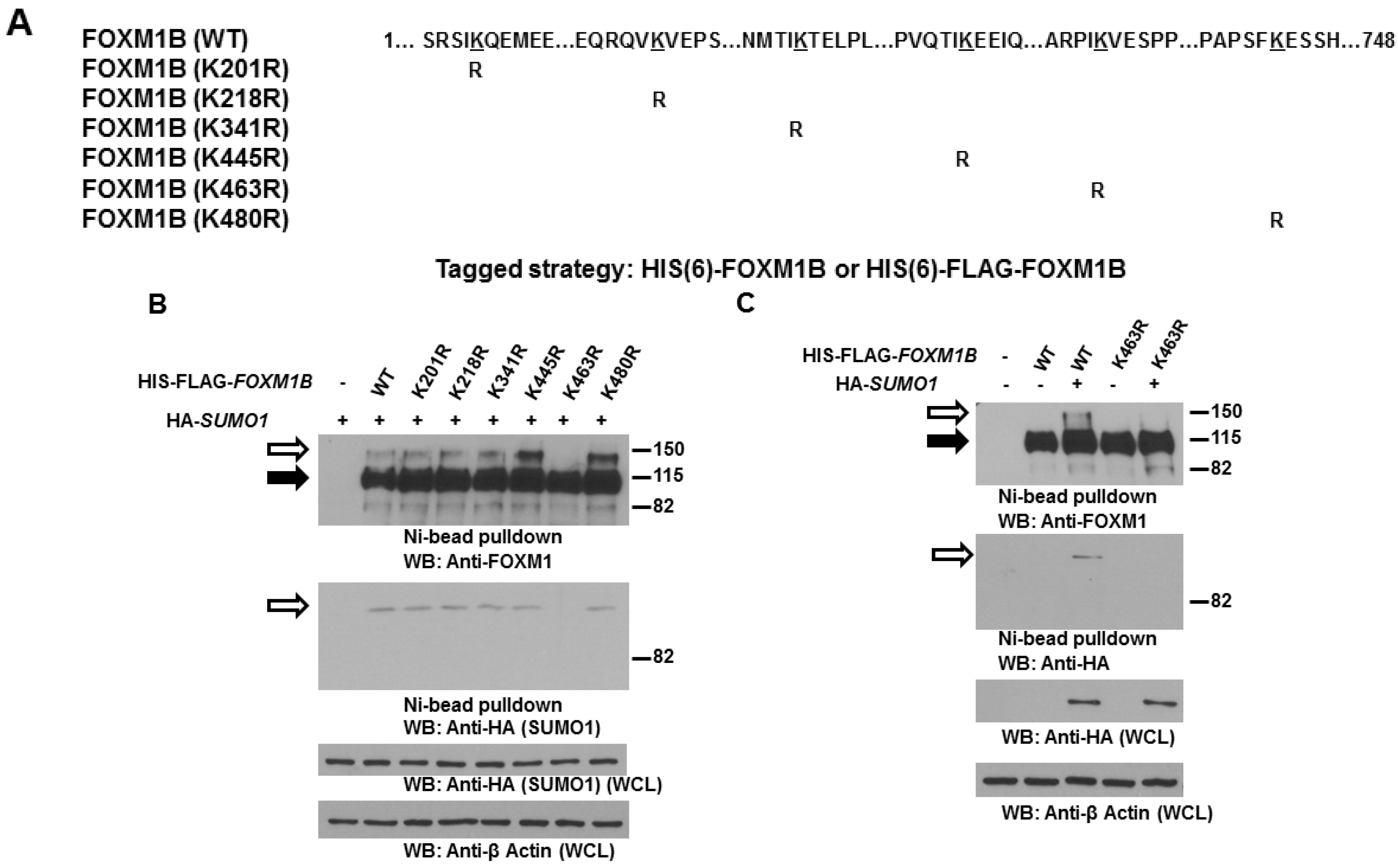
2.4. SUMOylation of FOXM1B at K463 Is Required for FOXM1B’s Transcriptional Activities
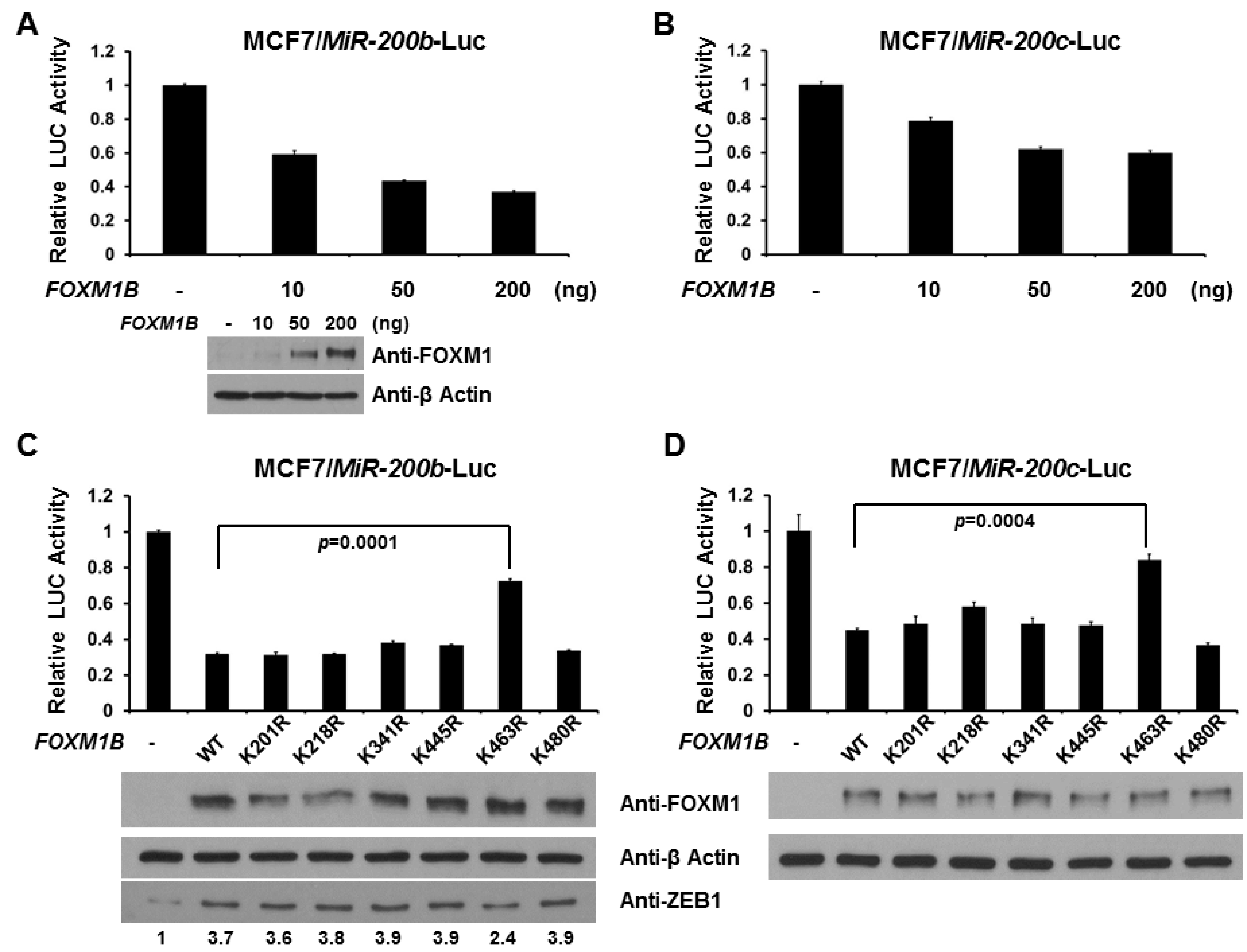
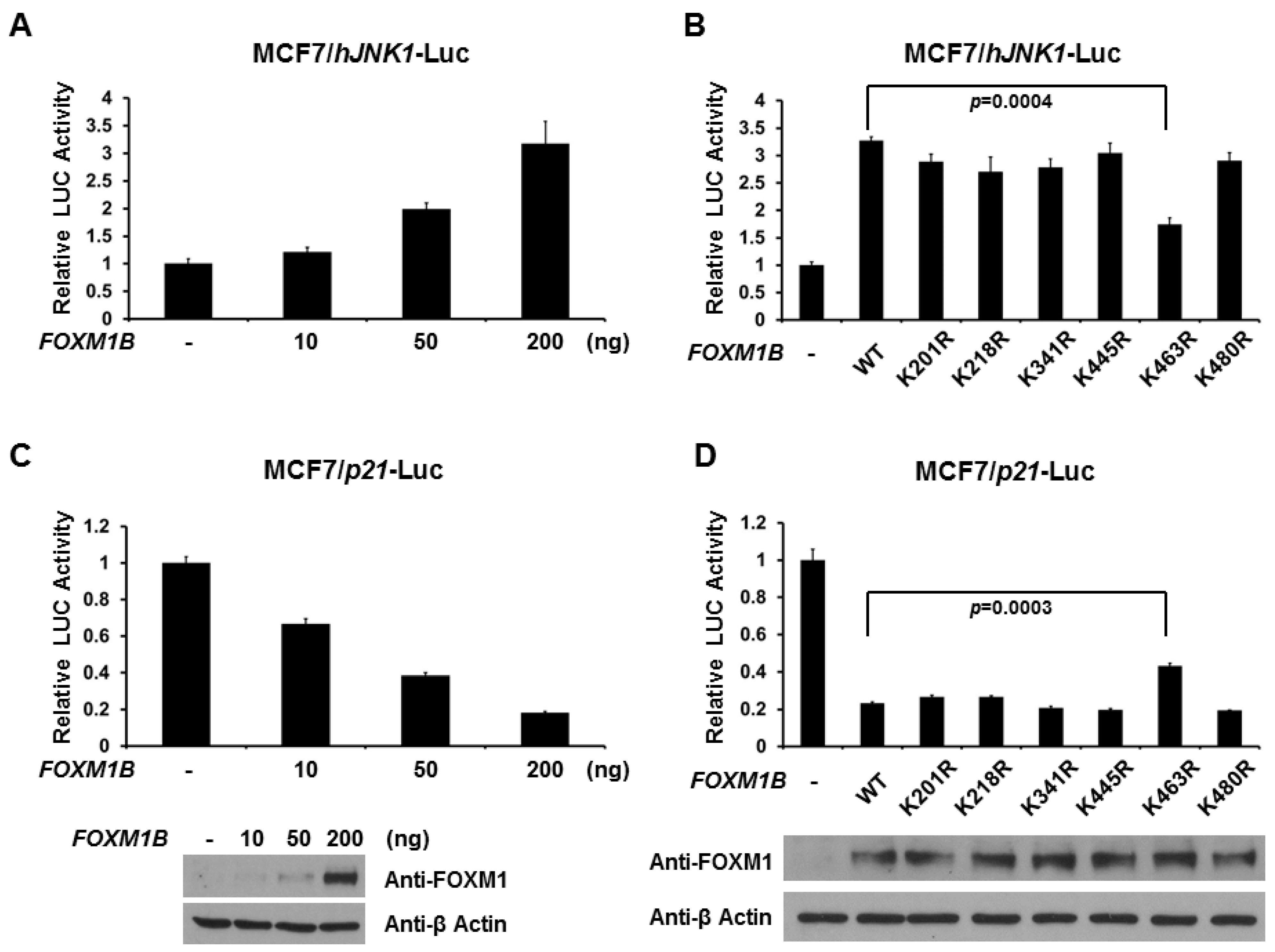
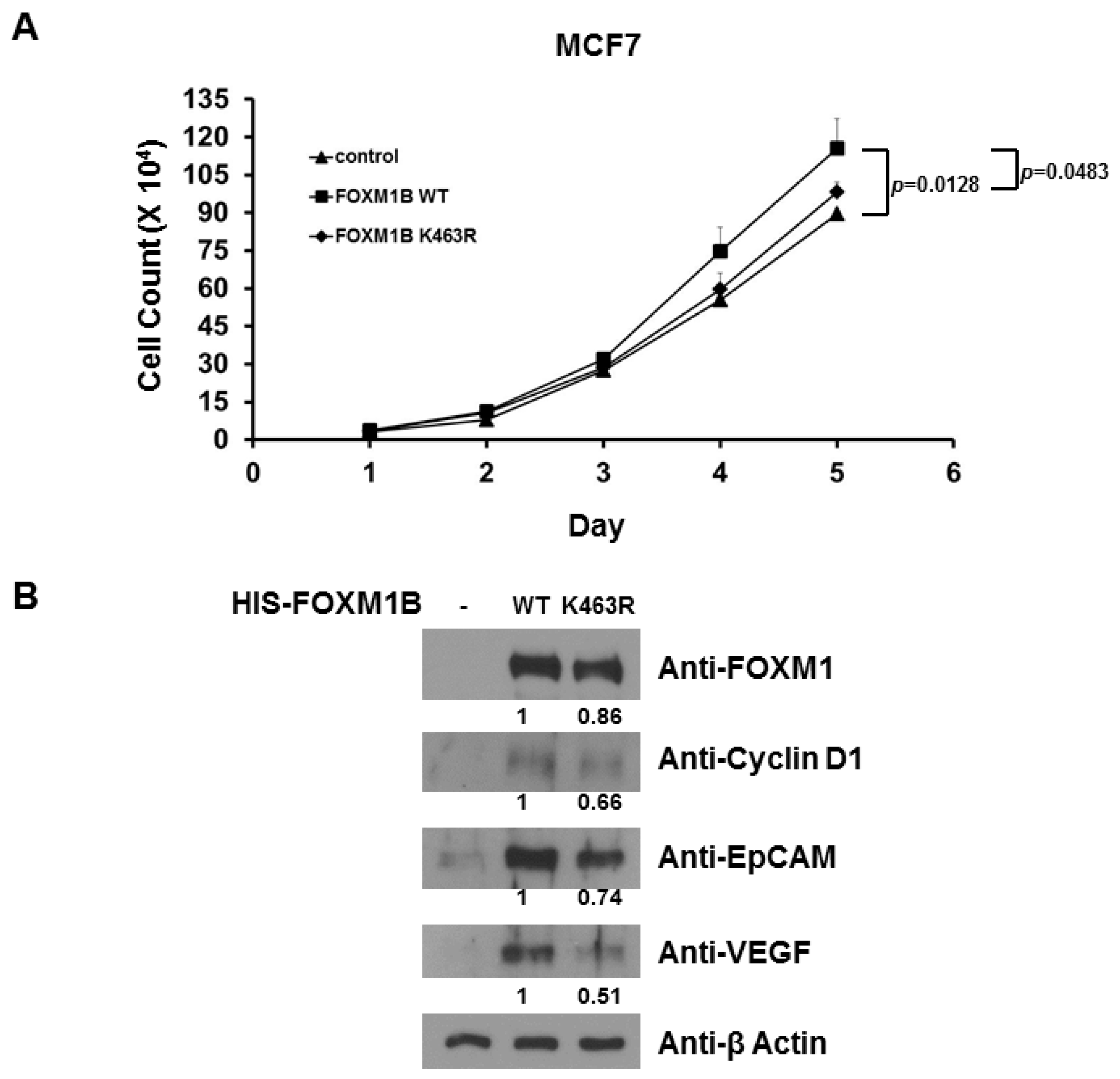
2.5. Loss of SUMOylation on FOXM1B Reduces Proliferation of MCF7 Cells
3. Discussion
4. Experimental Section
4.1. Reagents
4.2. DNA Constructs
4.3. Cell Culture and Transfection
4.4. Immunoprecipitation Assay
4.5. Immunoblotting
4.6. In Vivo SUMOylation Assays
4.7. Cell Proliferation Assay
4.8. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Abbreviations
Conflicts of Interest
References
- Raychaudhuri, P.; Park, H.J. FoxM1: A master regulator of tumor metastasis. Cancer Res. 2011, 71, 4329–4333. [Google Scholar] [CrossRef]
- Chen, X.; Müller, G.A.; Quaas, M.; Fischer, M.; Han, N.; Stutchbury, B.; Sharrocks, A.D.; Engeland, K. The forkhead transcription factor FOXM1 controls cell cycle-dependent gene expression through an atypical chromatin binding mechanism. Mol. Cell. Biol. 2013, 33, 227–236. [Google Scholar] [CrossRef]
- Nakamura, S.; Hirano, I.; Okinaka, K.; Takemura, T.; Yokota, D.; Ono, T.; Shigeno, K.; Shibata, K.; Fujisawa, S.; Ohnishi, K. The FOXM1 transcriptional factor promotes the proliferation of leukemia cells through modulation of cell cycle progression in acute myeloid leukemia. Carcinogenesis 2010, 31, 2012–2021. [Google Scholar] [CrossRef]
- Petrovic, V.; Costa, R.H.; Lau, L.F.; Raychaudhuri, P.; Tyner, A.L. FoxM1 regulates growth factor-induced expression of kinase-interacting stathmin (KIS) to promote cell cycle progression. J. Biol. Chem. 2008, 283, 453–460. [Google Scholar] [CrossRef]
- Qian, J.; Luo, Y.; Gu, X.; Zhan, W.; Wang, X. Twist1 promotes gastric cancer cell proliferation through up-regulation of FoxM1. PLoS One 2013, 8, e77625. [Google Scholar]
- Qu, K.; Xu, X.; Liu, C.; Wu, Q.; Wei, J.; Meng, F.; Zhou, L.; Wang, Z.; Lei, L.; Liu, P. Negative regulation of transcription factor FoxM1 by p53 enhances oxaliplatin-induced senescence in hepatocellular carcinoma. Cancer Lett. 2013, 331, 105–114. [Google Scholar] [CrossRef]
- Pandit, B.; Halasi, M.; Gartel, A.L. p53 negatively regulates expression of FoxM1. Cell Cycle 2009, 8, 3425–3427. [Google Scholar] [CrossRef]
- Barsotti, A.M.; Prives, C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene 2009, 28, 4295–4305. [Google Scholar] [CrossRef]
- Wierstra, I.; Alves, J. Transcription factor FOXM1c is repressed by RB and activated by cyclin D1/Cdk4. Biol. Chem. 2006, 387, 949–962. [Google Scholar]
- Kalinichenko, V.V.; Major, M.L.; Wang, X.; Petrovic, V.; Kuechle, J.; Yoder, H.M.; Dennewitz, M.B.; Shin, B.; Datta, A.; Raychaudhuri, P.; et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004, 18, 830–850. [Google Scholar] [CrossRef]
- Halasi, M.; Gartel, A.L. A novel mode of FoxM1 regulation: Positive auto-regulatory loop. Cell Cycle 2009, 8, 1966–1967. [Google Scholar] [CrossRef]
- Mirza, M.K.; Sun, Y.; Zhao, Y.D.; Potula, H.H.; Frey, R.S.; Vogel, S.M.; Malik, A.B.; Zhao, Y.Y. FoxM1 regulates re-annealing of endothelial adherens junctions through transcriptional control of β-catenin expression. J. Exp. Med. 2010, 207, 1675–1685. [Google Scholar] [CrossRef]
- Bowman, A.; Nusse, R. Location, location, location: FoxM1 mediates β-catenin nuclear translocation and promotes gliomatumorigenesis. Cancer Cell 2011, 20, 415–416. [Google Scholar] [CrossRef]
- Zhang, N.; Wei, P.; Gong, A.; Chiu, W.T.; Lee, H.T.; Colman, H.; Huang, H.; Xue, J.; Liu, M.; Wang, Y.; et al. FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell 2011, 20, 427–442. [Google Scholar] [CrossRef]
- Wang, X.; Hung, N.J.; Costa, R.H. Earlier expression of the transcription factor HFH-11B diminishes induction of p21 (CIP1/WAF1) levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology 2001, 33, 1404–1414. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, N.; Dai, B.; Liu, M.; Sawaya, R.; Xie, K.; Huang, S. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res. 2008, 68, 8733–8742. [Google Scholar] [CrossRef]
- Dai, B.; Kang, S.H.; Gong, W.; Liu, M.; Aldape, K.D.; Sawaya, R.; Huang, S. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene 2007, 26, 6212–6219. [Google Scholar] [CrossRef]
- Wang, I.C.; Chen, Y.J.; Hughes, D.E.; Ackerson, T.; Major, M.L.; Kalinichenko, V.V.; Costa, R.H.; Raychaudhuri, P.; Tyner, A.L.; Lau, L.F. FoxM1 regulates transcription of JNK1 to promote the G1/S transition and tumor cell invasiveness. J. Biol. Chem. 2008, 283, 20770–20778. [Google Scholar] [CrossRef]
- Waseem, A.; Ali, M.; Odell, E.W.; Fortune, F.; Teh, M.T. Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncol. 2010, 46, 536–542. [Google Scholar] [CrossRef] [Green Version]
- Wang, I.C.; Chen, Y.J.; Hughes, D.; Petrovic, V.; Major, M.L.; Park, H.J.; Tan, Y.; Ackerson, T.; Costa, R.H. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2–Cks1) ubiquitin ligase. Mol. Cell. Biol. 2005, 25, 10875–10894. [Google Scholar] [CrossRef]
- Madureira, P.A.; Varshochi, R.; Constantinidou, D.; Francis, R.E.; Coombes, R.C.; Yao, K.M.; Lam, E.W. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J. Biol. Chem. 2006, 281, 25167–25176. [Google Scholar] [CrossRef]
- Halasi, M.; Gartel, A.L. FOX (M1) news—It is cancer. Mol. Cancer Ther. 2013, 12, 245–254. [Google Scholar] [CrossRef]
- Xu, N.; Jia, D.; Chen, W.; Wang, H.; Liu, F.; Ge, H.; Zhu, X.; Song, Y.; Zhang, X.; Zhang, D.; et al. FoxM1 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor metastasis. PLoS One 2013, 8, e59412. [Google Scholar]
- Wu, X.R.; Chen, Y.H.; Liu, D.M.; Sha, J.J.; Xuan, H.Q.; Bo, J.J.; Huang, Y.R. Increased expression of forkhead box M1 protein is associated with poor prognosis in clear cell renal cell carcinoma. Med. Oncol. 2013, 30, 346. [Google Scholar] [CrossRef]
- Sun, H.C.; Li, M.; Lu, J.L.; Yan, D.W.; Zhou, C.Z.; Fan, J.W.; Qin, X.B.; Tang, H.M.; Peng, Z.H. Over-expression of Forkhead box M1 protein associates with aggressive tumor features and poor prognosis of hepatocellular carcinoma. Oncol. Rep. 2011, 25, 1533–1539. [Google Scholar]
- Kong, X.; Li, L.; Li, Z.; Le, X.; Huang, C.; Jia, Z.; Cui, J.; Huang, S.; Wang, L.; Xie, K. Dysregulated expression of FOXM1 isoforms drives progression of pancreatic cancer. Cancer Res. 2013, 73, 3987–3996. [Google Scholar] [CrossRef]
- Bao, B.; Wang, Z.; Ali, S.; Kong, D.; Banerjee, S.; Ahmad, A.; Li, Y.; Azmi, A.S.; Miele, L.; Sarkar, F.H. Over-expression of FoxM1 leads to epithelial-mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J. Cell Biochem. 2011, 112, 2296–2306. [Google Scholar] [CrossRef]
- Gong, L.; Ji, W.K.; Hu, X.H.; Hu, W.F.; Tang, X.C.; Huang, Z.X.; Li, L.; Liu, M.; Xiang, S.H.; Wu, E.; et al. Sumoylation differentially regulates Sp1 to control cell differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, 5574–5579. [Google Scholar] [CrossRef]
- Lee, G.Y.; Jang, H.; Lee, J.H.; Huh, J.Y.; Choi, S.; Chung, J.; Kim, J.B. PIASy-mediated sumoylation of SREBP1c regulates hepatic lipid metabolism upon fasting signaling. Mol. Cell. Biol. 2014, 34, 1720. [Google Scholar]
- Wang, C.M.; Brennan, V.C.; Gutierrez, N.M.; Wang, X.; Wang, L.; Yang, W.H. SUMOylation of ATF3 alters its transcriptional activity on regulation of TP53 gene. J. Cell Biochem. 2013, 114, 589–598. [Google Scholar] [CrossRef]
- Nepveu-Traversy, M.É.; Berthoux, L. The conserved sumoylation consensus site in TRIM5α modulates its immune activation functions. Virus Res. 2014, 184, 30–38. [Google Scholar] [CrossRef]
- Lee, P.C.; Taylor-Jaffe, K.M.; Nordin, K.M.; Prasad, M.S.; Lander, R.M.; LaBonne, C. SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J. Cell Biol. 2012, 198, 799–813. [Google Scholar] [CrossRef]
- Yang, W.H.; Heaton, J.H.; Brevig, H.; Mukherjee, S.; Iñiguez-Lluhí, J.A.; Hammer, G.D. SUMOylation inhibits SF-1 activity by reducing CDK7-mediated serine 203 phosphorylation. Mol. Cell. Biol. 2009, 29, 613–625. [Google Scholar] [CrossRef]
- Qin, Y.; Bao, H.; Pan, Y.; Yin, M.; Liu, Y.; Wu, S.; Li, H. SUMOylation alterations are associated with multidrug resistance in hepato cellular carcinoma. Mol. Med. Rep. 2014, 9, 877–881. [Google Scholar]
- Gareau, J.R.; Lima, C.D. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell. Biol. 2010, 11, 861–871. [Google Scholar] [CrossRef]
- Aukrust, I.; Bjørkhaug, L.; Negahdar, M.; Molnes, J.; Johansson, B.B.; Müller, Y.; Haas, W.; Gygi, S.P.; Søvik, O.; Flatmark, T.; et al. SUMOylation of pancreatic glucokinase regulates its cellular stability and activity. J. Biol. Chem. 2013, 288, 5951–5962. [Google Scholar] [CrossRef]
- Gong, Z.; Brackertz, M.; Renkawitz, R. SUMO modification enhances p66-mediated transcriptional repression of the Mi-2/NuRD complex. Mol. Cell. Biol. 2006, 26, 4519–4528. [Google Scholar] [CrossRef]
- Rytinki, M.M.; Palvimo, J.J. SUMOylation modulates the transcription repressor function of RIP140. J. Biol. Chem. 2008, 283, 11586–11595. [Google Scholar] [CrossRef]
- Abed, M.; Barry, K.C.; Kenyagin, D.; Koltun, B.; Phippen, T.M.; Delrow, J.J.; Parkhurst, S.M.; Orian, A. Degringolade, a SUMO-targeted ubiquitin ligase, inhibits Hairy/Groucho-mediated repression. EMBO J. 2011, 30, 1289–1301. [Google Scholar] [CrossRef]
- Duverger, O.; Chen, S.X.; Lee, D.; Li, T.; Chock, P.B.; Morasso, M.I. SUMOylation of DLX3 by SUMO1 promotes its transcriptional activity. J. Cell Biochem. 2011, 112, 445–452. [Google Scholar] [CrossRef]
- Rodriguez, M.S.; Desterro, J.M.; Lain, S.; Midgley, C.A.; Lane, D.P.; Hay, R.T. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999, 18, 6455–6461. [Google Scholar] [CrossRef]
- Guo, Y.; Yang, M.C.; Weissler, J.C.; Yang, Y.S. Modulation of PLAGL2 transactivation activity by Ubc9 co-activation not SUMOylation. Biochem. Biophys. Res. Commun. 2008, 374, 570–575. [Google Scholar] [CrossRef]
- Ihara, M.; Stein, P.; Schultz, R.M. UBE2I (UBC9), a SUMO-conjugating enzyme, localizes to nuclear speckles and stimulates transcription in mouse oocytes. Biol. Reprod. 2008, 79, 906–913. [Google Scholar] [CrossRef]
- Arora, T.; Liu, B.; He, H.; Kim, J.; Murphy, T.L.; Murphy, K.M.; Modlin, R.L.; Shuai, K. PIASx is a transcriptional co-repressor of signal transducer and activator of transcription 4. J. Biol. Chem. 2003, 278, 21327–213230. [Google Scholar]
- Joshi, K.; Banasavadi-Siddegowda, Y.; Mo, X.; Kim, S.H.; Mao, P.; Kig, C.; Nardini, D.; Sobol, R.W.; Chow, L.M.; Kornblum, H.I.; et al. MELK-dependent FOXM1 phosphorylation is essential for proliferation of glioma stem cells. Stem Cells 2013, 31, 1051–1063. [Google Scholar] [CrossRef]
- Chen, Y.J.; Dominguez-Brauer, C.; Wang, Z.; Asara, J.M.; Costa, R.H.; Tyner, A.L.; Lau, L.F.; Raychaudhuri, P. A conserved phosphorylation site within the forkhead domain of FoxM1B is required for its activation by cyclin-CDK1. J. Biol. Chem. 2009, 284, 30695–30707. [Google Scholar] [CrossRef]
- Fu, Z.; Malureanu, L.; Huang, J.; Wang, W.; Li, H.; van Deursen, J.M.; Tindall, D.J.; Chen, J. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat. Cell Biol. 2008, 10, 1076–1082. [Google Scholar] [CrossRef]
- Major, M.L.; Lepe, R.; Costa, R.H. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol. Cell. Biol. 2004, 24, 2649–2661. [Google Scholar] [CrossRef]
- Laoukili, J.; Stahl, M.; Medema, R.H. FoxM1: At the crossroads of ageing and cancer. Biochim. Biophys. Acta 2007, 1775, 92–102. [Google Scholar]
- Lam, A.K.; Ngan, A.W.; Leung, M.H.; Kwok, D.C.; Liu, V.W.; Chan, D.W.; Leung, W.Y.; Yao, K.M. FOXM1b, which is present at elevated levels in cancer cells, has a greater transforming potential than FOXM1c. Front. Oncol. 2013, 3, 11. [Google Scholar]
- Liu, M.; Dai, B.; Kang, S.H.; Ban, K.; Huang, F.J.; Lang, F.F.; Aldape, K.D.; Xie, T.X.; Pelloski, C.E.; Xie, K.; et al. FoxM1B is over-expressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006, 66, 3593–3602. [Google Scholar] [CrossRef]
- Kalinina, O.A.; Kalinin, S.A.; Polack, E.W.; Mikaelian, I.; Panda, S.; Costa, R.H.; Adami, G.R. Sustained hepatic expression of FoxM1B in transgenic mice has minimal effects on hepatocellular carcinoma development but increases cell proliferation rates in preneoplastic and early neoplastic lesions. Oncogene 2003, 22, 6266–6276. [Google Scholar] [CrossRef]
- Park, H.J.; Gusarova, G.; Wang, Z.; Carr, J.R.; Li, J.; Kim, K.H.; Qiu, J.; Park, Y.D.; Williamson, P.R.; Hay, N.; et al. Deregulation of FoxM1b leads to tumour metastasis. EMBO Mol. Med. 2011, 3, 21–34. [Google Scholar] [CrossRef]
- Knouf, E.C.; Garg, K.; Arroyo, J.D.; Correa, Y.; Sarkar, D.; Parkin, R.K.; Wurz, K.; O’Briant, K.C.; Godwin, A.K.; Urban, N.D.; et al. An integrative genomic approach identifies p73 and p63 as activators of miR-200 microRNA family transcription. Nucleic Acids Res. 2012, 40, 499–510. [Google Scholar]
- Bendoraite, A.; Knouf, E.C.; Garg, K.S.; Parkin, R.K.; Kroh, E.M.; O’Briant, K.C.; Ventura, A.P.; Godwin, A.K.; Karlan, B.Y.; Drescher, C.W.; et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: Evidence supporting a mesothelial-to-epithelial transition. Gynecol. Oncol. 2010, 116, 117–125. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Brabletz, S.; Brabletz, T. The ZEB/miR-200 feedback loop—A motor of cellular plasticity in development and cancer? EMBO Rep. 2010, 11, 670–677. [Google Scholar] [CrossRef]
- Myatt, S.S.; Kongsema, M.; Man, C.W.; Kelly, D.J.; Gomes, A.R.; Khongkow, P.; Karunarathna, U.; Zona, S.; Langer, J.K.; Dunsby, C.W.; et al. SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene 2014. [Google Scholar] [CrossRef]
- Schimmel, J.; Eifler, K.; Sigurðsson, J.O.; Cuijpers, S.A.; Hendriks, I.A.; Verlaan-de Vries, M.; Kelstrup, C.D; Francavilla, C.; Medema, R.H.; Olsen, J.V.; et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol. Cell 2014, 53, 1053–1066. [Google Scholar] [CrossRef]
- Dephoure, N.; Zhou, C.; Villén, J.; Beausoleil, S.A.; Bakalarski, C.E.; Elledge, S.J.; Gygi, S.P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 2008, 105, 10762–10767. [Google Scholar]
- Liu, R.; Wang, L.; Chen, G.; Katoh, H.; Chen, C.; Liu, Y.; Zheng, P. FOXP3 up-regulates p21 expression by site-specific inhibition of histone deacetylase 2/histone deacetylase 4 association to the locus. Cancer Res. 2009, 69, 2252–2259. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, C.-M.; Liu, R.; Wang, L.; Nascimento, L.; Brennan, V.C.; Yang, W.-H. SUMOylation of FOXM1B Alters Its Transcriptional Activity on Regulation of MiR-200 Family and JNK1 in MCF7 Human Breast Cancer Cells. Int. J. Mol. Sci. 2014, 15, 10233-10251. https://doi.org/10.3390/ijms150610233
Wang C-M, Liu R, Wang L, Nascimento L, Brennan VC, Yang W-H. SUMOylation of FOXM1B Alters Its Transcriptional Activity on Regulation of MiR-200 Family and JNK1 in MCF7 Human Breast Cancer Cells. International Journal of Molecular Sciences. 2014; 15(6):10233-10251. https://doi.org/10.3390/ijms150610233
Chicago/Turabian StyleWang, Chiung-Min, Runhua Liu, Lizhong Wang, Leticia Nascimento, Victoria C. Brennan, and Wei-Hsiung Yang. 2014. "SUMOylation of FOXM1B Alters Its Transcriptional Activity on Regulation of MiR-200 Family and JNK1 in MCF7 Human Breast Cancer Cells" International Journal of Molecular Sciences 15, no. 6: 10233-10251. https://doi.org/10.3390/ijms150610233




