Alternative Splicing in Plant Immunity
Abstract
:1. Introduction
2. Plant Disease Resistance
2.1. R Genes
2.2. Signaling Components in ETI
3. AS of R Genes
3.1. AS of TIR-NBS-LRR Genes
3.1.1. Arabidopsis RPS4
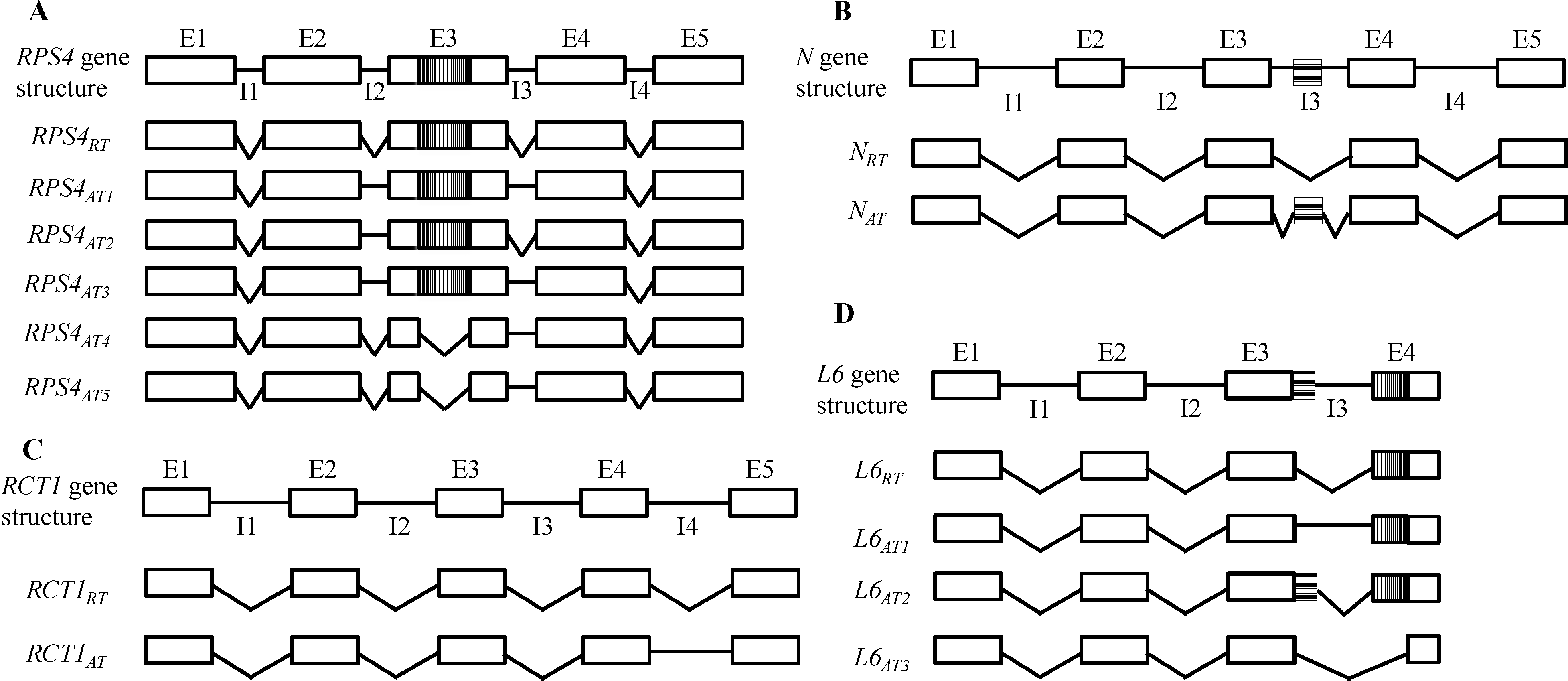
3.1.2. Tobacco N
3.1.3. M. truncatula RCT1
3.1.4. Flax L6 and Tomato Bs4
3.2. AS of CC-NBS-LRR Genes
4. Possible Mechanisms of AS-Mediated Regulation of Defense Response
4.1. Disruption of R Protein Autoinhibition
4.2. Function as Signaling Factors
5. Regulation of AS of R Genes
6. Future Prospects
Abbreviations
| ARC | apaf-1, R and ced-4 |
| Avr | avirulence |
| BCA2 | breast cancer-amplified sequence 2 |
| Bs4 | bacterial spot resistance gene 4 |
| CC | coil-coiled |
| Mla | mildew A |
| CDC5L | cell divison cycle 5 like protein |
| CPR1 | constitutive expresser of PR genes 1 |
| DC3000 | P. syringae pv tomato strain DC3000 |
| Dscam | down syndrome cell adhesion molecule |
| EDS1 | enhanced disease susceptibility 1 |
| EF-Tu | elongation factor-Tu |
| ETI | effector-triggered immunity |
| hnRNPs | heterogeneous nuclear ribonucleoproteins |
| HR | hypersensitive response |
| HRT | HR to TCV |
| LRR | leucine-rich repeat region |
| MAC | MOS4-associated complex |
| MOS | modifier of snc1 |
| NBS | nucleotide-binding site |
| NDR1 | non-race-specific disease resistance 1 |
| NMD | nonsense-mediated decay |
| NPR1 | non-expresser of PR genes 1 |
| p50 | 50-kDa helicase protein |
| PAD4 | phytoalexin deficient 4 |
| PAMPs or MAMPs | pathogen (or microbe)-associated molecular patterns |
| PLRG1 | pleiotropic regulator 1 |
| Prp19 | precursor RNA processing 19 |
| PRRs | pattern recognition receptors |
| PTI | PAMP-triggered immunity |
| RAC1 | resistance to Albugo candida isolated ACEM1 |
| RAN1 | RAS-related nuclear protein 1 |
| RBPs | RNA binding proteins |
| RGA | R-gene analog |
| RPP | recognition of Peronospora parasitica |
| RPS | resistance to P. Syringae |
| SCF | skp1-cullin1-F-box |
| snc1 | suppressor of npr1-1, constitutive 1 |
| snRNPs | small ribonucleoproteins |
| SR | serine-arginine rich |
| SRFR1 | suppressor of RPS4-RLD1 |
| TCV | turnip crinkle virus |
| TIR | Drosophila Toll and mammalian Interleukin-1 receptor |
| TMV | tobacco mosaic virus |
| TN | truncated R proteins containing TIR-NBS only |
| TX | truncated R proteins containing TIR only |
| uORFs | upstream ORFs |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nilsen, T.W.; Graveley, B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature 2010, 463, 457–463. [Google Scholar] [CrossRef]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Graveley, B.R. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell 2005, 123, 65–73. [Google Scholar] [CrossRef]
- Brett, D.; Pospisil, H.; Valcarcel, J.; Reich, J.; Bork, P. Alternative splicing and genome complexity. Nat. Genet. 2002, 30, 29–30. [Google Scholar] [CrossRef]
- Kazan, K. Alternative splicing and proteome diversity in plants: The tip of the iceberg has just emerged. Trends Plant Sci. 2003, 8, 468–471. [Google Scholar] [CrossRef]
- Ramani, A.K.; Calarco, J.A.; Pan, Q.; Mavandadi, S.; Wang, Y.; Nelson, A.C.; Lee, L.J.; Morris, Q.; Blencowe, B.J.; Zhen, M. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res. 2011, 21, 342–348. [Google Scholar] [CrossRef]
- Zhu, W.; Schlueter, S.D.; Brendel, V. Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol. 2003, 132, 469–484. [Google Scholar] [CrossRef]
- Iida, K.; Seki, M.; Sakurai, T.; Satou, M.; Akiyama, K.; Toyoda, T.; Konagaya, A.; Shinozaki, K. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 2004, 32, 5096–5103. [Google Scholar] [CrossRef]
- Wang, B.B.; Brendel, V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 2006, 103, 7175–7180. [Google Scholar] [CrossRef]
- Campbell, M.A.; Haas, B.J.; Hamilton, J.P.; Mount, S.M.; Buell, C.R. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BioMed Cent. Genomics 2006, 7, 327. [Google Scholar]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef]
- Ali, G.S.; Reddy, A.S. Regulation of alternative splicing of pre-mRNAs by stresses. Curr. Top. Microbiol. 2008, 326, 257–275. [Google Scholar]
- Mastrangelo, A.M.; Marone, D.; Laido, G.; de Leonardis, A.M.; de Vita, P. Alternative splicing: Enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012, 185–186, 40–49. [Google Scholar] [CrossRef]
- Howard, B.E.; Hu, Q.; Babaoglu, A.C.; Chandra, M.; Borghi, M.; Tan, X.; He, L.; Winter-Sederoff, H.; Gassmann, W.; Veronese, P. High-throughput RNA sequencing of pseudomonas-infected Arabidopsis reveals hidden transcriptome complexity and novel splice variants. PLoS One 2013, 8, e74183. [Google Scholar]
- Lopato, S.; Kalyna, M.; Dorner, S.; Kobayashi, R.; Krainer, A.R.; Barta, A. AtSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 1999, 13, 987–1001. [Google Scholar] [CrossRef]
- Lopato, S.; Waigmann, E.; Barta, A. Characterization of a novel arginine/serine-rich splicing factor in Arabidopsis. Plant Cell 1996, 8, 2255–2264. [Google Scholar] [CrossRef]
- Loraine, A.E.; McCormick, S.; Estrada, A.; Patel, K.; Qin, P. RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 2013, 162, 1092–1109. [Google Scholar] [CrossRef]
- Yoshimura, K.; Yabuta, Y.; Ishikawa, T.; Shigeoka, S. Identification of a cis element for tissue-specific alternative splicing of chloroplast ascorbate peroxidase pre-mRNA in higher plants. J. Biol. Chem. 2002, 277, 40623–40632. [Google Scholar] [CrossRef]
- De la Fuente van Bentem, S.; Vossen, J.H.; Vermeer, J.E.; de Vroomen, M.J.; Gadella, T.W., Jr.; Haring, M.A.; Cornelissen, B.J. The subcellular localization of plant protein phosphatase 5 isoforms is determined by alternative splicing. Plant Physiol. 2003, 133, 702–712. [Google Scholar] [CrossRef]
- Kriechbaumer, V.; Wang, P.; Hawes, C.; Abell, B.M. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 2012, 70, 292–302. [Google Scholar] [CrossRef]
- Remy, E.; Cabrito, T.R.; Baster, P.; Batista, R.A.; Teixeira, M.C.; Friml, J.; Sa-Correia, I.; Duque, P. A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 2013, 25, 901–926. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, S.D.; Saraiva, R.; Maia, T.M.; Abreu, I.A.; Duque, P. XBAT35, a novel Arabidopsis RING E3 ligase exhibiting dual targeting of its splice isoforms, is involved in ethylene-mediated regulation of apical hook curvature. Mol. Plant 2012, 5, 1295–1309. [Google Scholar] [CrossRef]
- Reddy, A.S.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef]
- Carvalho, R.F.; Feijao, C.V.; Duque, P. On the physiological significance of alternative splicing events in higher plants. Protoplasma 2013, 250, 639–650. [Google Scholar] [CrossRef]
- Reddy, A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef]
- Staiger, D.; Brown, J.W.S. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef]
- Zhang, X.C.; Gassmann, W. RPS4-mediated disease resistance requires the combined presence of RPS4 transcripts with full-length and truncated open reading frames. Plant Cell 2003, 15, 2333–2342. [Google Scholar] [CrossRef]
- Zhang, X.C.; Gassmann, W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 2007, 145, 1577–1587. [Google Scholar] [CrossRef]
- Dinesh-Kumar, S.P.; Baker, B.J. Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 2000, 97, 1908–1913. [Google Scholar] [CrossRef]
- Tang, F.; Yang, S.; Gao, M.; Zhu, H. Alternative splicing is required for RCT1-mediated disease resistance in Medicago truncatula. Plant Mol. Biol. 2013, 82, 367–374. [Google Scholar] [CrossRef]
- Staiger, D.; Korneli, C.; Lummer, M.; Navarro, L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2013, 197, 394–404. [Google Scholar] [CrossRef]
- Dodds, P.N.; Lawrence, G.J.; Catanzariti, A.M.; Teh, T.; Wang, C.I.; Ayliffe, M.A.; Kobe, B.; Ellis, J.G. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 8888–8893. [Google Scholar] [CrossRef]
- Van der Biezen, E.A.; Jones, J.D. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 1998, 23, 454–456. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Macho, A.P.; Schwessinger, B.; Ntoukakis, V.; Brutus, A.; Segonzac, C.; Roy, S.; Kadota, Y.; Oh, M.H.; Sklenar, J.; Derbyshire, P. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 2014, 343, 1509–1512. [Google Scholar] [CrossRef]
- Kim, S.H.; Kwon, S.I.; Saha, D.; Anyanwu, N.C.; Gassmann, W. Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol. 2009, 150, 1723–1732. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Halane, M.K.; Kim, S.H.; Gassmann, W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 2011, 334, 1405–1408. [Google Scholar] [CrossRef]
- Glazebrook, J.; Rogers, E.E.; Ausubel, F.M. Use of Arabidopsis for genetic dissection of plant defense responses. Annu. Rev. Genet. 1997, 31, 547–569. [Google Scholar] [CrossRef]
- Goodman, R.N.; Novacky, A.J. The Hypersensitive Reaction in Plants to Pathogens: A Resistance Phenomenon; APS Press: St. Paul, MN, USA, 1994. [Google Scholar]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Shen, Q.H.; Saijo, Y.; Mauch, S.; Biskup, C.; Bieri, S.; Keller, B.; Seki, H.; Ulker, B.; Somssich, I.E.; Schulze-Lefert, P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007, 315, 1098–1103. [Google Scholar] [CrossRef]
- Traut, T.W. The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur. J. Biochem. 1994, 222, 9–19. [Google Scholar] [CrossRef]
- Rairdan, G.J.; Moffett, P. Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 2006, 18, 2082–2093. [Google Scholar] [CrossRef]
- Kobe, B.; Deisenhofer, J. The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 1994, 19, 415–421. [Google Scholar] [CrossRef]
- Ellis, J.G.; Lawrence, G.J.; Luck, J.E.; Dodds, P.N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 1999, 11, 495–506. [Google Scholar] [CrossRef]
- Leister, R.T.; Katagiri, F. A resistance gene product of the nucleotide binding site—Leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J. 2000, 22, 345–354. [Google Scholar] [CrossRef]
- Jia, Y.; McAdams, S.A.; Bryan, G.T.; Hershey, H.P.; Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. Embo J. 2000, 19, 4004–4014. [Google Scholar] [CrossRef]
- Tao, Y.; Yuan, F.; Leister, R.T.; Ausubel, F.M.; Katagiri, F. Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 2000, 12, 2541–2554. [Google Scholar]
- Boyes, D.C.; Nam, J.; Dangl, J.L. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 1998, 95, 15849–15854. [Google Scholar] [CrossRef]
- Aarts, N.; Metz, M.; Holub, E.; Staskawicz, B.J.; Daniels, M.J.; Parker, J.E. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 10306–10311. [Google Scholar]
- Chandra-Shekara, A.C.; Navarre, D.; Kachroo, A.; Kang, H.G.; Klessig, D.; Kachroo, P. Signaling requirements and role of salicylic acid in HRT- and rrt-mediated resistance to turnip crinkle virus in Arabidopsis. Plant J. 2004, 40, 647–659. [Google Scholar] [CrossRef]
- Bittner-Eddy, P.D.; Beynon, J.L. The Arabidopsis downy mildew resistance gene, RPP13-Nd, functions independently of NDR1 and EDS1 and does not require the accumulation of salicylic acid. Mol. Plant Microbe Interact. 2001, 14, 416–421. [Google Scholar] [CrossRef]
- McDowell, J.M.; Dhandaydham, M.; Long, T.A.; Aarts, M.G.; Goff, S.; Holub, E.B.; Dangl, J.L. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 1998, 10, 1861–1874. [Google Scholar] [CrossRef]
- Venugopal, S.C.; Jeong, R.D.; Mandal, M.K.; Zhu, S.; Chandra-Shekara, A.C.; Xia, Y.; Hersh, M.; Stromberg, A.J.; Navarre, D.; Kachroo, A. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genet. 2009, 5, e1000545. [Google Scholar] [CrossRef]
- Falk, A.; Feys, B.J.; Frost, L.N.; Jones, J.D.; Daniels, M.J.; Parker, J.E. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 1999, 96, 3292–3297. [Google Scholar] [CrossRef]
- Feys, B.J.; Wiermer, M.; Bhat, R.A.; Moisan, L.J.; Medina-Escobar, N.; Neu, C.; Cabral, A.; Parker, J.E. Arabidopsis senescence-associated gene101 stabilizes and signals within an enhanced disease susceptibility1 complex in plant innate immunity. Plant Cell 2005, 17, 2601–2613. [Google Scholar]
- Zhu, S.; Jeong, R.D.; Venugopal, S.C.; Lapchyk, L.; Navarre, D.; Kachroo, A.; Kachroo, P. SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog. 2011, 7, e1002318. [Google Scholar] [CrossRef]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef]
- Li, X.; Clarke, J.D.; Zhang, Y.; Dong, X. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 2001, 14, 1131–1139. [Google Scholar] [CrossRef]
- Palma, K.; Zhang, Y.; Li, X. An importin α homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 2005, 15, 1129–1135. [Google Scholar]
- Palma, K.; Zhao, Q.; Cheng, Y.T.; Bi, D.; Monaghan, J.; Cheng, W.; Zhang, Y.; Li, X. Regulation of plant innate immunity by three proteins in a complex conserved across the plant and animal kingdoms. Genes Dev. 2007, 21, 1484–1493. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, X. A putative nucleoporin 96 is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell 2005, 17, 1306–1316. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, Y.T.; Bi, D.; Palma, K.; Li, X. MOS2, a protein containing G-patch and KOW motifs, is essential for innate immunity in Arabidopsis thaliana. Curr. Biol. 2005, 15, 1936–1942. [Google Scholar] [CrossRef]
- Goritschnig, S.; Zhang, Y.; Li, X. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 2007, 49, 540–551. [Google Scholar] [CrossRef]
- Wiermer, M.; Palma, K.; Zhang, Y.; Li, X. Should I stay or should I go? Nucleocytoplasmic trafficking in plant innate immunity. Cell. Microbiol. 2007, 9, 1880–1890. [Google Scholar] [CrossRef]
- Goritschnig, S.; Weihmann, T.; Zhang, Y.; Fobert, P.; McCourt, P.; Li, X. A novel role for protein farnesylation in plant innate immunity. Plant Physiol. 2008, 148, 348–357. [Google Scholar] [CrossRef]
- Cheng, Y.T.; Germain, H.; Wiermer, M.; Bi, D.; Xu, F.; Garcia, A.V.; Wirthmueller, L.; Despres, C.; Parker, J.E.; Zhang, Y. Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 2009, 21, 2503–2516. [Google Scholar] [CrossRef]
- Germain, H.; Qu, N.; Cheng, Y.T.; Lee, E.; Huang, Y.; Dong, O.X.; Gannon, P.; Huang, S.; Ding, P.; Li, Y. MOS11: A new component in the mRNA export pathway. PLoS Genet. 2010, 6, e1001250. [Google Scholar] [CrossRef]
- Li, Y.; Tessaro, M.J.; Li, X.; Zhang, Y. Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol. 2010, 153, 1425–1434. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, Z.; Jing, B.; Gannon, P.; Ding, J.; Xu, F.; Li, X.; Zhang, Y. Transportin-SR is required for proper splicing of resistance genes and plant immunity. PLoS Genet. 2011, 7, e1002159. [Google Scholar] [CrossRef]
- Xu, F.; Xu, S.; Wiermer, M.; Zhang, Y.; Li, X. The cyclin L homolog MOS12 and the MOS4-associated complex are required for the proper splicing of plant resistance genes. Plant J. 2012, 70, 916–928. [Google Scholar] [CrossRef]
- Whitham, S.; Dinesh-Kumar, S.P.; Choi, D.; Hehl, R.; Corr, C.; Baker, B. The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 1994, 78, 1101–1115. [Google Scholar] [CrossRef]
- Ayliffe, M.A.; Frost, D.V.; Finnegan, E.J.; Lawrence, G.J.; Anderson, P.A.; Ellis, J.G. Analysis of alternative transcripts of the flax L6 rust resistance gene. Plant J. 1999, 17, 287–292. [Google Scholar] [CrossRef]
- Borhan, M.H.; Holub, E.B.; Beynon, J.L.; Rozwadowski, K.; Rimmer, S.R. The Arabidopsis TIR-NB-LRR gene RAC1 confers resistance to Albugo candida (white rust) and is dependent on EDS1 but not PAD4. Mol. Plant Microbe Interact. 2004, 17, 711–719. [Google Scholar] [CrossRef]
- Parker, J.E.; Coleman, M.J.; Szabo, V.; Frost, L.N.; Schmidt, R.; van der Biezen, E.A.; Moores, T.; Dean, C.; Daniels, M.J.; Jones, J.D. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 1997, 9, 879–894. [Google Scholar] [CrossRef]
- Gassmann, W.; Hinsch, M.E.; Staskawicz, B.J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999, 20, 265–277. [Google Scholar] [CrossRef]
- Yi, H.; Richards, E.J. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 2007, 19, 2929–2939. [Google Scholar] [CrossRef]
- Schornack, S.; Ballvora, A.; Gurlebeck, D.; Peart, J.; Baulcombe, D.; Ganal, M.; Baker, B.; Bonas, U.; Lahaye, T. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 2004, 37, 46–60. [Google Scholar] [CrossRef]
- Vidal, S.; Cabrera, H.; Andersson, R.A.; Fredriksson, A.; Valkonen, J.P. Potato gene Y-1 is an N gene homolog that confers cell death upon infection with potato virus Y. Mol. Plant Microbe Interact. 2002, 15, 717–727. [Google Scholar] [CrossRef]
- Yang, S.; Gao, M.; Xu, C.; Gao, J.; Deshpande, S.; Lin, S.; Roe, B.A.; Zhu, H. Alfalfa benefits from Medicago truncatula: The RCT1 gene from M. truncatula confers broad-spectrum resistance to anthracnose in alfalfa. Proc. Natl. Acad. Sci. USA 2008, 105, 12164–12169. [Google Scholar] [CrossRef]
- Erickson, F.L.; Holzberg, S.; Calderon-Urrea, A.; Handley, V.; Axtell, M.; Corr, C.; Baker, B. The helicase domain of the TMV replicase proteins induces the N-mediated defence response in tobacco. Plant J. 1999, 18, 67–75. [Google Scholar] [CrossRef]
- Bendahmane, A.; Querci, M.; Kanyuka, K.; Baulcombe, D.C. Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 2000, 21, 73–81. [Google Scholar] [CrossRef]
- Van der Vossen, E.A.; van der Voort, J.N.; Kanyuka, K.; Bendahmane, A.; Sandbrink, H.; Baulcombe, D.C.; Bakker, J.; Stiekema, W.J.; Klein-Lankhorst, R.M. Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: A virus and a nematode. Plant J. 2000, 23, 567–576. [Google Scholar] [CrossRef]
- Moffett, P.; Farnham, G.; Peart, J.; Baulcombe, D.C. Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. Embo J. 2002, 21, 4511–4519. [Google Scholar] [CrossRef]
- Gassmann, W. Alternative splicing in plant defense. Curr. Top. Microbiol. 2008, 326, 219–233. [Google Scholar]
- Nandety, R.S.; Caplan, J.L.; Cavanaugh, K.; Perroud, B.; Wroblewski, T.; Michelmore, R.W.; Meyers, B.C. The role of TIR-NBS and TIR-X proteins in plant basal defense responses. Plant Physiol. 2013, 162, 1459–1472. [Google Scholar]
- Frost, D.; Way, H.; Howles, P.; Luck, J.; Manners, J.; Hardham, A.; Finnegan, J.; Ellis, J. Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol. Plant Microbe Interact. 2004, 17, 224–232. [Google Scholar] [CrossRef]
- Bernoux, M.; Ve, T.; Williams, S.; Warren, C.; Hatters, D.; Valkov, E.; Zhang, X.; Ellis, J.G.; Kobe, B.; Dodds, P.N. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011, 9, 200–211. [Google Scholar] [CrossRef]
- Sela, H.; Spiridon, L.N.; Petrescu, A.J.; Akerman, M.; Mandel-Gutfreund, Y.; Nevo, E.; Loutre, C.; Keller, B.; Schulman, A.H.; Fahima, T. Ancient diversity of splicing motifs and protein surfaces in the wild emmer wheat (Triticum dicoccoides) LR10 coiled coil (CC) and leucine-rich repeat (LRR) domains. Mol. Plant. Pathol. 2012, 13, 276–287. [Google Scholar] [CrossRef] [Green Version]
- Saintenac, C.; Zhang, W.; Salcedo, A.; Rouse, M.N.; Trick, H.N.; Akhunov, E.; Dubcovsky, J. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 2013, 341, 783–786. [Google Scholar] [CrossRef]
- Halterman, D.A.; Wei, F.; Wise, R.P. Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 2003, 131, 558–567. [Google Scholar] [CrossRef]
- Halterman, D.; Zhou, F.; Wei, F.; Wise, R.P.; Schulze-Lefert, P. The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 2001, 25, 335–348. [Google Scholar] [CrossRef]
- Costanzo, S.; Jia, Y.L. Alternatively spliced transcripts of Pi-ta blast resistance gene in Oryza sativa. Plant Sci. 2009, 177, 468–478. [Google Scholar] [CrossRef]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef]
- Ferrier-Cana, E.; Macadre, C.; Sevignac, M.; David, P.; Langin, T.; Geffroy, V. Distinct post-transcriptional modifications result into seven alternative transcripts of the CC-NBS-LRR gene JA1tr of Phaseolus vulgaris. Theor. Appl. Genet. 2005, 110, 895–905. [Google Scholar] [CrossRef]
- Okuyama, Y.; Kanzaki, H.; Abe, A.; Yoshida, K.; Tamiru, M.; Saitoh, H.; Fujibe, T.; Matsumura, H.; Shenton, M.; Galam, D.C. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011, 66, 467–479. [Google Scholar] [CrossRef]
- Halterman, D.A.; Wise, R.P. Upstream open reading frames of the barley Mla13 powdery mildew resistance gene function co-operatively to down-regulate translation. Mol. Plant. Pathol. 2006, 7, 167–176. [Google Scholar] [CrossRef]
- Lukasik, E.; Takken, F.L. STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 2009, 12, 427–436. [Google Scholar] [CrossRef]
- Bendahmane, A.; Farnham, G.; Moffett, P.; Baulcombe, D.C. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 2002, 32, 195–204. [Google Scholar] [CrossRef]
- Shirano, Y.; Kachroo, P.; Shah, J.; Klessig, D.F. A gain-of-function mutation in an Arabidopsis toll interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 2002, 14, 3149–3162. [Google Scholar] [CrossRef]
- Zhang, Y.; Goritschnig, S.; Dong, X.; Li, X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 2003, 15, 2636–2646. [Google Scholar] [CrossRef]
- Belkhadir, Y.; Subramaniam, R.; Dangl, J.L. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 2004, 7, 391–399. [Google Scholar] [CrossRef]
- Maekawa, T.; Cheng, W.; Spiridon, L.N.; Toller, A.; Lukasik, E.; Saijo, Y.; Liu, P.; Shen, Q.H.; Micluta, M.A.; Somssich, I.E. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011, 9, 187–199. [Google Scholar] [CrossRef]
- Takken, F.L.; Goverse, A. How to build a pathogen detector: Structural basis of NB-LRR function. Curr. Opin. Plant Biol. 2012, 15, 375–384. [Google Scholar] [CrossRef]
- Rairdan, G.J.; Collier, S.M.; Sacco, M.A.; Baldwin, T.T.; Boettrich, T.; Moffett, P. The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 2008, 20, 739–751. [Google Scholar] [CrossRef]
- Qi, D.; deYoung, B.J.; Innes, R.W. Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol. 2012, 158, 1819–1832. [Google Scholar] [CrossRef]
- Mackey, D.; Belkhadir, Y.; Alonso, J.M.; Ecker, J.R.; Dangl, J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef]
- Mackey, D.; Holt, B.F., III; Wiig, A.; Dangl, J.L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 2002, 108, 743–754. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Bi, D.; Cheng, Y.T.; Li, X.; Zhang, Y. SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 2010, 6, e1001111. [Google Scholar] [CrossRef]
- Cheng, Y.T.; Li, Y.; Huang, S.; Huang, Y.; Dong, X.; Zhang, Y.; Li, X. Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc. Natl. Acad. Sci. USA 2011, 108, 14694–14699. [Google Scholar] [CrossRef]
- Zhang, Y.; Dorey, S.; Swiderski, M.; Jones, J.D. Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 2004, 40, 213–224. [Google Scholar] [CrossRef]
- Collier, S.M.; Hamel, L.P.; Moffett, P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 2011, 24, 918–931. [Google Scholar] [CrossRef]
- Swiderski, M.R.; Birker, D.; Jones, J.D.G. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant Microbe Interact. 2009, 22, 157–165. [Google Scholar] [CrossRef]
- Meyers, B.C.; Morgante, M.; Michelmore, R.W. TIR-X and TIR-NBS proteins: Two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 2002, 32, 77–92. [Google Scholar] [CrossRef]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Burch-Smith, T.M.; Schiff, M.; Caplan, J.L.; Tsao, J.; Czymmek, K.; Dinesh-Kumar, S.P. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007, 5, e68. [Google Scholar] [CrossRef]
- Wirthmueller, L.; Zhang, Y.; Jones, J.D.; Parker, J.E. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 2007, 17, 2023–2029. [Google Scholar] [CrossRef]
- Heidrich, K.; Wirthmueller, L.; Tasset, C.; Pouzet, C.; Deslandes, L.; Parker, J.E. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science 2011, 334, 1401–1404. [Google Scholar] [CrossRef]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 1999, 11, 781–792. [Google Scholar] [CrossRef]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef]
- Gassmann, W. Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol. Plant Microbe Interact. 2005, 18, 1054–1060. [Google Scholar] [CrossRef]
- Chen, M.; Manley, J.L. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009, 10, 741–754. [Google Scholar]
- Day, I.S.; Golovkin, M.; Palusa, S.G.; Link, A.; Ali, G.S.; Thomas, J.; Richardson, D.N.; Reddy, A.S. Interactions of SR45, an SR-like protein, with spliceosomal proteins and an intronic sequence: Insights into regulated splicing. Plant J. 2012, 71, 936–947. [Google Scholar] [CrossRef]
- Thomas, J.; Palusa, S.G.; Prasad, K.V.; Ali, G.S.; Surabhi, G.K.; Ben-Hur, A.; Abdel-Ghany, S.E.; Reddy, A.S. Identification of an intronic splicing regulatory element involved in auto-regulation of alternative splicing of SCL33 pre-mRNA. Plant J. 2012. [Google Scholar] [CrossRef]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007, 49, 1091–1107. [Google Scholar] [CrossRef]
- Ajuh, P.; Sleeman, J.; Chusainow, J.; Lamond, A.I. A direct interaction between the C-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J. Biol. Chem. 2001, 276, 42370–42381. [Google Scholar] [CrossRef]
- Zhou, Z.; Licklider, L.J.; Gygi, S.P.; Reed, R. Comprehensive proteomic analysis of the human spliceosome. Nature 2002, 419, 182–185. [Google Scholar] [CrossRef]
- Monaghan, J.; Xu, F.; Gao, M.; Zhao, Q.; Palma, K.; Long, C.; Chen, S.; Zhang, Y.; Li, X. Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 2009, 5, e1000526. [Google Scholar]
- Monaghan, J.; Xu, F.; Xu, S.; Zhang, Y.; Li, X. Two putative RNA-binding proteins function with unequal genetic redundancy in the MOS4-associated complex. Plant Physiol. 2010, 154, 1783–1793. [Google Scholar] [CrossRef]
- Caplan, J.L.; Mamillapalli, P.; Burch-Smith, T.M.; Czymmek, K.; Dinesh-Kumar, S.P. Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 2008, 132, 449–462. [Google Scholar]
- Maquat, L.E. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 2004, 5, 89–99. [Google Scholar] [CrossRef]
- McGlincy, N.J.; Smith, C.W. Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem. Sci. 2008, 33, 385–393. [Google Scholar] [CrossRef]
- Jeong, H.J.; Kim, Y.J.; Kim, S.H.; Kim, Y.H.; Lee, I.J.; Kim, Y.K.; Shin, J.S. Nonsense-mediated mRNA decay factors, UPF1 and UPF3, contribute to plant defense. Plant Cell Physiol. 2011, 52, 2147–2156. [Google Scholar] [CrossRef]
- Rayson, S.; Arciga-Reyes, L.; Wootton, L.; de Torres Zabala, M.; Truman, W.; Graham, N.; Grant, M.; Davies, B. A role for nonsense-mediated mRNA decay in plants: Pathogen responses are induced in Arabidopsis thaliana NMD mutants. PLoS One 2012, 7, e31917. [Google Scholar]
- Kalyna, M.; Simpson, C.G.; Syed, N.H.; Lewandowska, D.; Marquez, Y.; Kusenda, B.; Marshall, J.; Fuller, J.; Cardle, L.; McNicol, J. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012, 40, 2454–2469. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Yang, S.; Tang, F.; Zhu, H. Alternative Splicing in Plant Immunity. Int. J. Mol. Sci. 2014, 15, 10424-10445. https://doi.org/10.3390/ijms150610424
Yang S, Tang F, Zhu H. Alternative Splicing in Plant Immunity. International Journal of Molecular Sciences. 2014; 15(6):10424-10445. https://doi.org/10.3390/ijms150610424
Chicago/Turabian StyleYang, Shengming, Fang Tang, and Hongyan Zhu. 2014. "Alternative Splicing in Plant Immunity" International Journal of Molecular Sciences 15, no. 6: 10424-10445. https://doi.org/10.3390/ijms150610424




