Mass Spectrometry Methodology in Lipid Analysis
Abstract
:1. Introduction
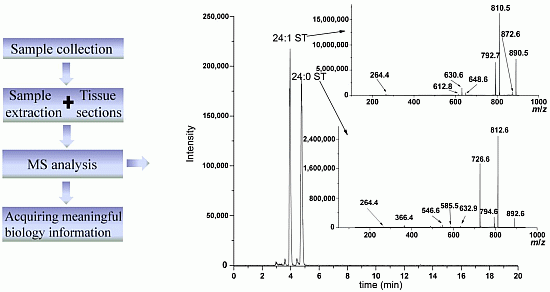
2. The Methods for Lipid Extraction
3. The Ionization Technologies of MS
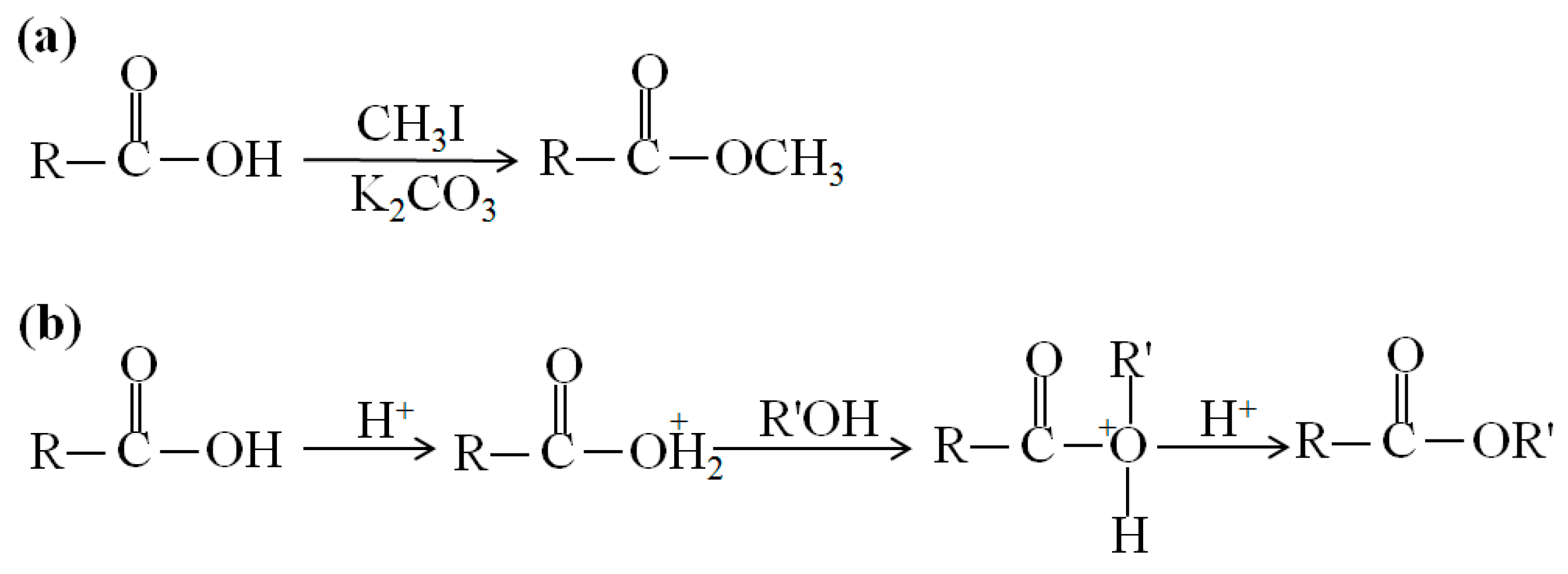
3.1. Electron Ionization (EI) and Chemical Ionization (CI)
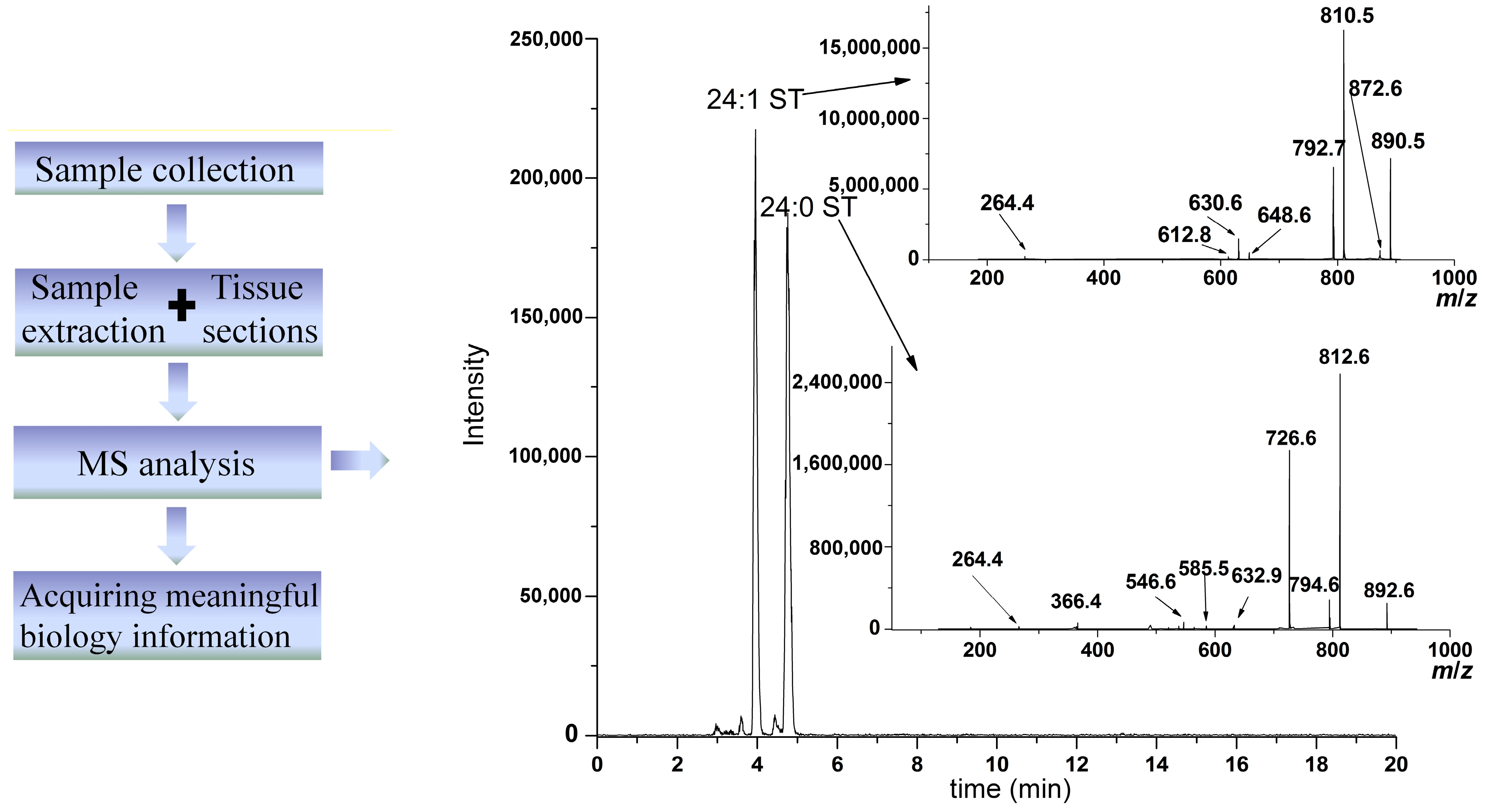
3.2. Fast Atom Bombardment (FAB)
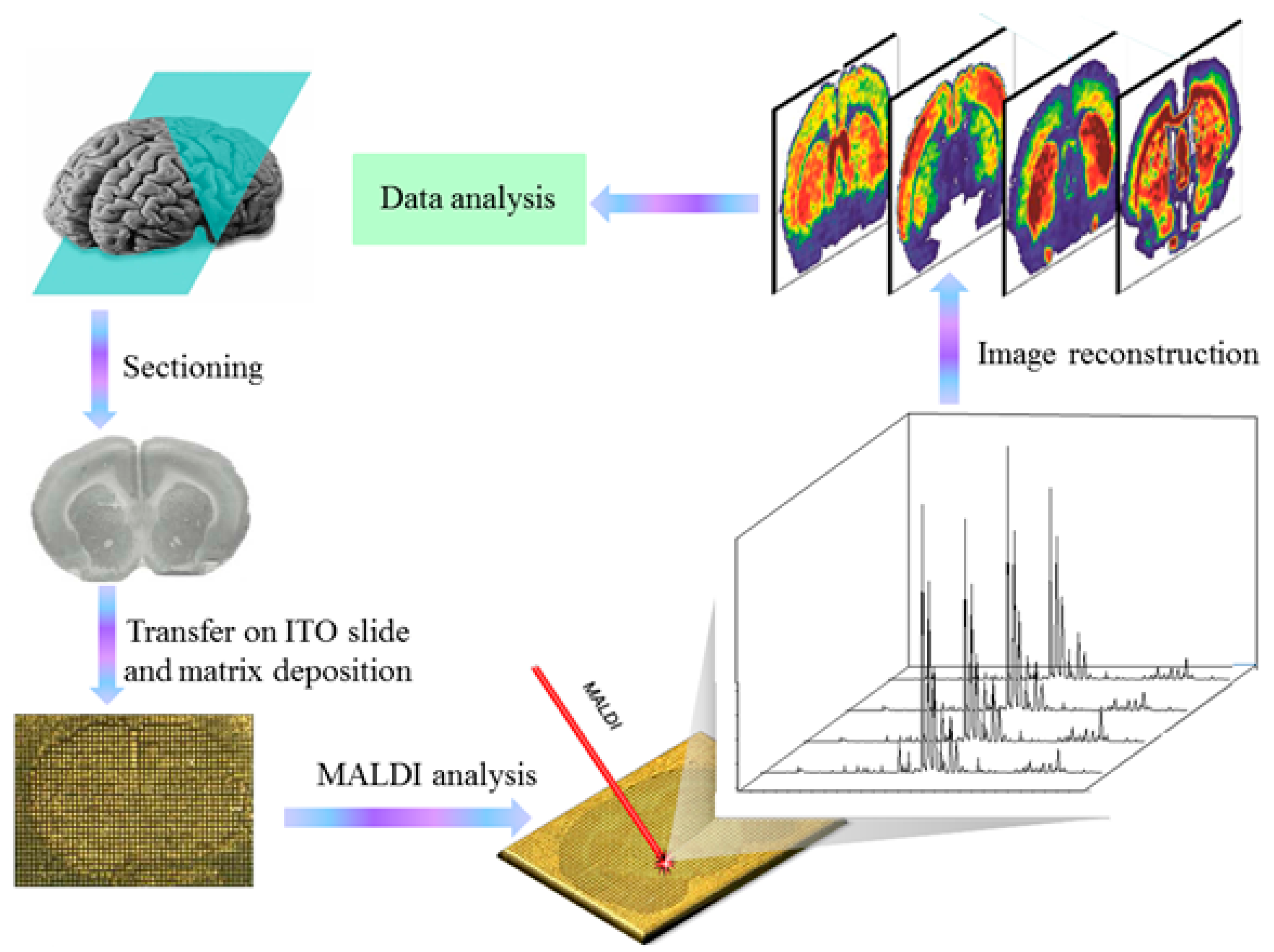
3.3. Matrix-Assisted Laser Desorption Ionization (MALDI)
3.4. Electrospray Ionization (ESI), Atmosphere Pressure Chemical Ionization (APCI), Atmosphere Pressure Photoionization (APPI) and Desorption Electrospray Ionization (DESI)
4. The Mass Analyzers of MS
5. The Bioinformatics Technology for Data Processing
6. Outlooks and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Han, X.L.; Gross, R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: A bridge to lipidomics. J. Lipid Res. 2003, 44, 1071–1079. [Google Scholar] [CrossRef]
- Li, F.; Prestwich, G.D. Functional Lipidomics; CRC Press: Boca Raton, FL, USA, 2005; p. 2. [Google Scholar]
- Shevchenko, A.; Simons, K. Lipidomics: Coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010, 11, 593–598. [Google Scholar] [CrossRef]
- Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2010, 29, 112–124. [Google Scholar]
- Lam, S.M.; Shui, G. Lipidomics as a principal tool for advancing biomedical research. J. Genet. Genomics 2013, 40, 375–390. [Google Scholar] [CrossRef]
- Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef]
- Fuchs, B.; Süß, R.; Teubera, K.; Eibisch, M.; Schiller, J. Lipid analysis by thin-layer chromatography—A review of the current state. J. Chromatogr. A 2011, 1218, 2754–2774. [Google Scholar] [CrossRef]
- Malins, D.C.; Mangold, H.K. Analysis of complex lipid mixtures by thin-layer chromatography and complementary methods. J. Am. Oil Chem. Soc. 1960, 37, 576–578. [Google Scholar] [CrossRef]
- Ramstedt, B.; Leppimaki, P.; Axberg, M.; Slotte, J.P. Analysis of natural and synthetic sphingomyelins using high-performance thin-layer chromatography. Eur. J. Biochem. 1999, 266, 997–1002. [Google Scholar] [CrossRef]
- Tang, B.; Row, K.H. Development of gas chromatography analysis of fatty acids in marine organisms. J. Chromatogr. Sci. 2013, 51, 599–607. [Google Scholar] [CrossRef]
- Volin, P. Analysis of steroidal lipids by gas and liquid chromatography. J. Chromatogr. A 2001, 935, 125–140. [Google Scholar] [CrossRef]
- Yang, Z.; Parrish, C.C.; Helleur, R.J. Automated gas chromatographic method for neutral lipid carbon number profiles in marine samples. J. Chromatogr. Sci. 1996, 34, 556–568. [Google Scholar] [CrossRef]
- Saeed, S.; Howell, N.K. High-performance liquid chromatography and spectroscopic studies on fish oil oxidation products extracted from frozen atlantic mackerel. J. Am. Oil Chem. Soc. 1999, 76, 391–397. [Google Scholar] [CrossRef]
- Goodridge, C.F.; Beaudry, R.M.; Pestka, J.J.; Smith, D.M. ELISA for monitoring lipid oxidation in chicken myofibrils through quantification of hexanal-protein adducts. J. Agric. Food Chem. 2003, 17, 7533–7539. [Google Scholar]
- Igarashi, T.; Aursand, M.; Hirata, Y.; Gribbestad, I.S.; Wada, S.; Nonaka, M. Nondestructive quantitative determination of docosahexaenoic acid and n-3 fatty acids in fish oils by high-resolution 1H nuclear magnetic resonance spectroscopy. J. Am. Oil Chem. Soc. 2003, 77, 737–748. [Google Scholar]
- Knothe, G.; Kenar, J.A. Determination of the fatty acid profile by 1H NMR spectroscopy. Eur. J. Lipid Sci. Technol. 2004, 106, 88–96. [Google Scholar] [CrossRef]
- Harkewicz, R.; Dennis, E.A. Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 2011, 80, 301–325. [Google Scholar] [CrossRef]
- Welti, R.; Wang, X.M. Lipid species profiling: A high-throughput approach to identifylipid compositional changes and determine the function of genes involved in lipid metabolism and signaling. Curr. Opin. Plant Biol. 2004, 7, 337–344. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Yatomi, Y.; Ohmori, T.; Rile, G.; Kazama, F.; Okamoto, H.; Sano, T.; Satoh, K.; Kume, S.; Tigyi, G.; Igarashi, Y. Sphingosine-1-phosphate as a major bioactive lysophospholipid that is released from platelets and interacts with endothelial cells. Blood 2000, 96, 3431–3438. [Google Scholar]
- Scherer, M.; Schmitz, G.; Liebisch, G. High-throughput analysis of sphingosine 1-phosphate, sphinganine 1-phosphate, and lysophosphatidic acid in plasma samples by liquid chromatography-tandem mass spectrometry. Clin. Chem. 2009, 55, 1218–1222. [Google Scholar] [CrossRef]
- Murph, M.; Tanaka, T.; Peng, J.; Felix, E.; Liu, S.; Trost, R.; Godwin, A.K.; Newman, R.; Mills, G. Liquid chromatography mass spectrometry for quantifying plasma lysophospholipids: Potential biomarkers for cancer diagnosis. Methods Enzymol. 2007, 433, 1–25. [Google Scholar] [CrossRef]
- Baker, D.L.; Morrison, P.; Miller, B.; Riely, C.A.; Tolley, B.; Westermann, A.M.; Bonfrer, J.M.; Bais, E.; Moolenaar, W.H.; Tigyi, G. Plasma lysophosphatidic acid concentration and ovarian cancer. J. Am. Med. Assoc. 2002, 287, 3081–3082. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1346. [Google Scholar] [CrossRef]
- Zhao, Z.W.; Xu, Y. An extremely simple method for extraction of lysophospholipids and phospholipids from blood samples. J. Lipid Res. 2010, 51, 652–659. [Google Scholar] [CrossRef]
- Reis, A.; Rudnitskaya, A.; Blackburn, G.J.; Mohd-Fauzi, N.; Pitt, A.R.; Spickett, C.M. A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J. Lipid Res. 2013, 54, 1812–1824. [Google Scholar] [CrossRef]
- Quehenberger, O.; Armando, A.M.; Dennis, E.A. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim. Biophys. Acta 2011, 1811, 648–656. [Google Scholar] [CrossRef]
- Morishige, J.; Urikura, M.; Takagi, H.; Hirano, K.; Koike, T.; Tanaka, T.; Satouchi, K. A clean-up technology for the simultaneous determination of lysophosphatidic acid and sphingosine-1-phosphate by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry using a phosphate-capture molecule, Phos-tag. Rapid Commun. Mass Spectrom. 2010, 24, 1075–1084. [Google Scholar] [CrossRef]
- Narayanaswamy, P.; Shinde, S.; Sulc, R.; Kraut, R.; Staples, G.; Thiam, C.H.; Grimm, R.; Sellergren, B.; Torta, F.; Wenk, M.R. Lipidomic “Deep Profiling”: An enhanced workflow to reveal new molecular species of signaling lipids. Anal. Chem. 2014, 86, 3043–3047. [Google Scholar] [CrossRef]
- Ruiz, J.; Antequera, T.; Andres, A.I.; Petron, M.J.; Muriel, E. Improvement of a solid phase extraction method for analysis of lipid fractions in muscle foods. Anal. Chim. Acta 2004, 520, 201–205. [Google Scholar] [CrossRef]
- Newman, A.E.; Chin, E.H.; Schmidt, K.L.; Bond, L.; Wynne-Edwards, K.E.; Soma, K.K. Analysis of steroids in songbird plasma and brain by coupling solid phase extraction to radioimmunoassay. Gen. Comp. Endocrinol. 2008, 155, 503–510. [Google Scholar] [CrossRef]
- Firl, N.; Kienberger, H.; Hauser, T.; Rychlik, M. Determination of the fatty acid profile of neutral lipids, free fatty acids and phospholipids inhuman plasma. Clin. Chem. Lab. Med. 2013, 51, 799–810. [Google Scholar]
- Tulipani, S.; Llorach, R.; Urpi-Sarda, M.; Andres-Lacueva, C. Comparative analysis of sample preparation methods to handle the complexity of the blood fluid metabolome: When less is more. Anal. Chem. 2013, 85, 341–348. [Google Scholar] [CrossRef]
- Hoffman, E.; Stroobant, V. Mass Spectrometry: Principles and Applications, 3rd ed.; John Wiley and Son: Chichester, WS, UK, 2007. [Google Scholar]
- Denkert, C.; Budczies, J.; Kind, T.; Weichert, W.; Tablack, P.; Sehouli, J.; Niesporek, S.; Könsgen, D.; Dietel, M.; Fiehn, O. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 2006, 66, 10795–10804. [Google Scholar] [CrossRef]
- Ahmida, H.S.; Bertucci, P.; Franzo, L.; Massoud, R.; Cortese, C.; Lala, A.; Federici, G. Simultaneous determination of plasmatic phytosterols and cholesterol precursors using gas chromatography-mass spectrometry (GC-MS) with selective ion monitoring (SIM). J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2006, 842, 43–47. [Google Scholar] [CrossRef]
- Son, H.H.; Moon, J.Y.; Seo, H.S.; Kim, H.H.; Chung, B.C.; Choi, M.H. High-temperature GC-MS-based serum cholesterol signatures may reveal sex differences in vasospastic angina. J. Lipid Res. 2014, 55, 155–162. [Google Scholar] [CrossRef]
- Lin, Y.H.; Salem, N., Jr.; Wells, E.M.; Zhou, W.; Loewke, J.D.; Brown, J.A.; Lands, W.E.; Goldman, L.R.; Hibbeln, J.R. Automated high-throughput fatty acid analysis of umbilical cord serum and application to an epidemiological study. Lipids 2012, 47, 527–539. [Google Scholar] [CrossRef]
- Barkawi, L.S.; Cohen, J.D. A method for concurrent diazomethane synthesis and substrate methylation in a 96-sample format. Nat. Protoc. 2010, 5, 1619–1626. [Google Scholar] [CrossRef]
- Ji, H.; Voinov, V.G.; Deinzer, M.L.; Barofsky, D.F. Distinguishing between cis/trans isomers of monounsaturated fatty acids by FAB MS. Anal. Chem. 2007, 79, 1519–1522. [Google Scholar] [CrossRef]
- Gil, J.H.; Hong, J.Y.; Jung, J.H.; Kim, K.J.; Hong, J. Structural determination of monoacylglycerols extracted from marine sponge by fast atom bombardment tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007, 21, 1264–1270. [Google Scholar] [CrossRef]
- Hong, J.; Kim, Y.H.; Gil, J.H.; Cho, K.; Jung, J.H.; Han, S.Y. Structural determination of hexadecanoic lysophosphatidylcholine regioisomers by fast atom bombardment tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2002, 16, 2089–2093. [Google Scholar]
- Korachi, M.; Blinkhorn, A.S.; Drucker, D.B. Analysis of phospholipid molecular species distributions by fast atom bombardment mass spectrometry (FAB-MS). Eur. J. Lipid Sci. Technol. 2002, 104, 50–56. [Google Scholar] [CrossRef]
- Ohashi, Y.; Tanaka, T.; Akashi, S.; Morimoto, S.; Kishimoto, Y.; Nagai, Y. Squid nerve sphingomyelin containing an unusual sphingoid base. J. Lipid Res. 2000, 41, 1118–1124. [Google Scholar]
- Ahn, Y.M.; Lee, W.W.; Jung, J.H.; Lee, S.G.; Hong, J. Structural determination of glucosylceramides isolated from marine sponge by fast atom bombardment collision-induced dissociation linked scan at constant B/E. J. Mass Spectrom. 2009, 44, 1698–1708. [Google Scholar]
- Hoyer, T.; Tuszynski, W.; Lienau, C. Ultrafast photodimerization dynamics in α-cyano-4-hydroxycinnamic and sinapinic acid crystals. Chem. Phys. Lett. 2007, 443, 107–112. [Google Scholar] [CrossRef]
- Fuchs, B.; Schiller, J. Recent developments of useful MALDI matrices for the mass spectrometric characterization of apolar compounds. Curr. Org. Chem. 2009, 13, 1664–1681. [Google Scholar]
- Fujita, T.; Fujino, T.; Hirabayashi, K.; Korenaga, T. MALDI mass spectrometry using 2,4,6-trihydroxyacetophenone and 2,4-dihydroxyacetophenone with cyclodextrins: Suppression of matrix-related ions in low-molecular-weight region. Anal. Sci. 2010, 26, 743–748. [Google Scholar] [CrossRef]
- Steven, R.T.; Race, A.M.; Bunch, J. para-Nitroaniline is a promising matrix for MALDI-MS imaging on intermediate pressure MS systems. J. Am. Soc. Mass Spectrom. 2013, 24, 801–804. [Google Scholar] [CrossRef]
- Fuchs, B.; Bischoff, A.; Süss, R.; Teuber, K.; Schürenberg, M.; Suckau, D.; Schiller, J. Phosphatidylcholines and -ethanolamines can be easily mistaken in phospholipid mixtures: A negative ion MALDI-TOF MS study with 9-aminoacridine as matrix and egg yolk as selected example. Anal. Bioanal. Chem. 2009, 395, 2479–2487. [Google Scholar] [CrossRef]
- Bonnel, D.; Franck, J.; Mériaux, C.; Salzet, M.; Fournier, I. Ionic matrices pre-spotted matrix-assisted laser desorption/ionization plates for patient maker following in course of treatment, drug titration, and MALDI mass spectrometry imaging. Anal. Biochem. 2013, 434, 187–198. [Google Scholar] [CrossRef]
- McAlpin, C.R.; Voorhees, K.J.; Corpuz, A.R.; Richards, R.M. Analysis of lipids: Metal oxide laser ionization mass spectrometry. Anal. Chem. 2012, 84, 7677–7683. [Google Scholar] [CrossRef]
- Son, J.; Lee, G.; Cha, S. Direct analysis of triacylglycerols from crude lipid mixtures by gold nanoparticle-assisted laser desorption/ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 2014, 25, 891–894. [Google Scholar] [CrossRef]
- Wei, Y.B.; Li, S.M.; Wang, J.X.; Shu, C.Y.; Liu, J.A.; Xiong, S.X.; Song, J.W.; Zhang, J.J.; Zhao, Z.W. Polystyrene spheres-assisted matrix-assisted laser desorption ionization mass spectrometry for quantitative analysis of plasma lysophosphatidylcholines. Anal. Chem. 2013, 85, 4729–4734. [Google Scholar] [CrossRef]
- Ida, C.G.; Giuseppe, A.; Jais, J.P.; Sands, D.; Nowakowska, A.; Colas, J.; Sermet-Gaudelus, I.; Schuerenberg, M.; Piomelli, D.; Edelman, A. A novel lipidomic strategy reveals plasma phospholipid signatures associated with respiratory disease severity in cystic fibrosis patients. PLoS One 2009, 4, e7735. [Google Scholar] [CrossRef]
- Franck, J.; Arafah, K.; Elayed, M.; Bonnel, D.; Vergara, D.; Jacquet, A.; Vinatier, D.; Wisztorski, M.; Day, R.; Fournier, I. MALDI imaging mass spectrometry. Mol. Cell. Proteomics 2009, 8, 2023–2033. [Google Scholar] [CrossRef]
- Griffiths, R.L.; Sarsby, J.; Guggenheim, E.J.; Race, A.M.; Steven, R.T.; Fear, J.; Lalor, P.F.; Bunch, J. Formal lithium fixation improves direct analysis of lipids in tissue by mass spectrometry. Anal. Chem. 2013, 85, 7146–7153. [Google Scholar] [CrossRef]
- Longuespée, R.; Boyon, C.; Desmons, A.; Kerdraon, O.; Leblanc, E.; Farré, I.; Vinatier, D.; Day, R.; Fournier, I.; Salzet, M. Spectroimmunohistochemistry: A novel form of MALDI mass spectrometry imaging coupled to immunohistochemistry for tracking antibodies. OMICS 2014, 18, 132–141. [Google Scholar] [CrossRef]
- Hirano, H.; Masaki, N.; Hayasaka, T.; Watanabe, Y.; Masumoto, K.; Nagata, T.; Katou, F.; Setou, M. Matrix-assisted laser desorption/ionization imaging mass spectrometry revealed traces of dental problem associated with dental structure. Anal. Bioanal. Chem. 2014, 406, 1355–1363. [Google Scholar] [CrossRef]
- Goto, T.; Terada, N.; Inoue, T.; Nakayama, K.; Okada, Y.; Yoshikawa, T.; Miyazaki, Y.; Uegaki, M.; Sumiyoshi, S.; Kobayashi, T. ashi, T. The expression profile of phosphatidylinositol in high spatial resolution imaging mass spectrometry as a potential biomarker for prostat cancer. PLoS One. 2014, 28, e90242. [Google Scholar]
- Schober, Y.; Guenther, S.; Spengler, B.; Römpp, A. Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal. Chem. 2012, 84, 6293–6297. [Google Scholar] [CrossRef]
- Barnaby, O.; Wa, C.; Cerny, R.L.; Clarke, W.; Hage, D.S. Quantitative analysis of human Serum albumin using 16O/18O-labeling and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin. Chim. Acta 2010, 411, 1102–1110. [Google Scholar] [CrossRef]
- Ye, H.P.; Hill, J.; Kauffman, J.; Han, X.L. Qualitative and quantitative comparison of brand name and generic protein pharmaceuticals using isotope tags for relative and absolute quantification and matrix-assisted laser desorption/ionization tandem time-of-flight mass spectrometry. Anal. Biochem. 2010, 400, 46–55. [Google Scholar] [CrossRef]
- Han, X.L.; Gross, R.W. Shotgun lipidomics: Electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005, 24, 367–412. [Google Scholar] [CrossRef]
- Han, X.L.; Rozen, S.; Boyle, S.H.; Hellegers, C.; Cheng, H.; Burke, J.R.; Welsh-Bohmer, K.A.; Doraiswamy, P.M.; Kaddurah-Daouk, R. Metabolomics in early Alzheimer’s disease: Identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS One 2011, 6, e21643. [Google Scholar] [CrossRef]
- Zhao, Z.W.; Xu, Y. Measurement of endogenous lysophosphatidic acid by ESI-MS/MS in plasma samples requires pre-separation of lysophosphatidylcholine. J. Chromatogr. B 2009, 877, 3739–3742. [Google Scholar] [CrossRef]
- Sterz, K.; Scherer, G.; Ecker, J. A simple and robust UPLC-SRM/MS method to quantify urinary eicosanoids. J. Lipid Res. 2012, 53, 1026–1036. [Google Scholar] [CrossRef]
- Wang, S.Y.; Li, J.; Shi, X.Z.; Qiao, L.Z.; Lu, X.; Xu, G.W. A novel stop-flow two-dimensional liquid chromatography-mass spectrometry method for lipid analysis. J. Chromatogr. A 2013, 1321, 65–72. [Google Scholar] [CrossRef]
- Chen, S.; Yin, P.; Zhao, X.; Xing, W.; Hu, C.; Zhou, L.; Xu, G.W. Serum lipid profiling of patients with chronic hepatitis B, cirrhosis, and hepatocellular carcinoma by ultra-fast LC/IT-TOF MS. Electrophoresis 2013, 34, 2848–2856. [Google Scholar]
- Gao, F.; Zhang, Z.X.; Fu, X.F.; Li, W.; Wang, T.; Liu, H.W. Analysis of phospholipids by NACE with on-line ESI-MS. Electrophoresis 2007, 28, 1418–1425. [Google Scholar] [CrossRef]
- Sun, T.; Pawlowski, S.; Johnson, M.E. Highly efficient microscale purification of glycerophospholipids by microfluidic cell lysis and lipid extraction for lipidomics profiling. Anal. Chem. 2011, 83, 6628–6634. [Google Scholar] [CrossRef]
- Donota, F.; Cazalsc, G.; Gunataa, Z.; Egronc, D.; Malingeb, J.; Struba, C.; Fontanaa, A.; Schorr-Galindo, S. Analysis of neutral lipids from microalgae by HPLC-ELSD and APCI-MS/MS. J. Chromatogr. B 2013, 98–106. [Google Scholar]
- Mei, H.; Hsieh, Y.S.; Nardo, C.; Xu, X.Y.; Wang, S.Y.; Ng, K.; Korfmacher, W.A. Investigation of matrix effects in bioanalytical high-performance liquid chromatography/tandem mass spectrometric assays: Application to drug discovery. Rapid Commun. Mass Spectrom. 2003, 17, 97–103. [Google Scholar]
- Hanold, K.A.; Fischer, S.M.; Cormia, P.H.; Miller, C.E.; Syage, J.A. Atmospheric pressure photoionization 1: General properties for LC/MS. Anal.Chem. 2004, 76, 2842–2851. [Google Scholar] [CrossRef]
- Tian, H.; Bai, J.; An, Z.; Chen, Y.; Zhang, R.; He, J.; Bi, X.; Song, Y.; Abliz, Z. Plasma metabolome analysis by integrated ionization rapid-resolution liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 2071–2080. [Google Scholar] [CrossRef]
- Takáts, Z.; Wiseman, J.M.; Gologan, B.; Cooks, R.G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 2004, 306, 471–473. [Google Scholar] [CrossRef]
- Eberlin, L.S.; Norton, I.; Dill, A.L.; Golby, A.J.; Ligon, K.L.; Santagata, S.; Cooks, R.G.; Agar, N.Y. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 2012, 72, 645–654. [Google Scholar] [CrossRef]
- Abbassi-Ghadi, N.; Veselkov, K.; Kumar, S.; Huang, J.; Jones, E.; Strittmatter, N.; Kudo, H.; Goldin, R.; Takáts, Z.; Hanna, G.B. Discrimination of lymph node metastases using desorption electrospray ionisation-mass spectrometry imaging. Chem. Commun. 2014, 50, 3661–3664. [Google Scholar] [CrossRef]
- Thomas, M.; Mitchell, T.; Harman, D.; Deeley, J.; Nealon, J.; Blanksby, S. Ozone-induced dissociation: Elucidation of double bond position within mass-selected lipid ions. Anal. Chem. 2008, 80, 303–311. [Google Scholar]
- Thomas, M.; Mitchell, T.; Harman, D.; Deeley, J.; Murphy, R.; Blanksby, S. Elucidation of double bond position in unsaturated lipids by ozone electrospray ionization mass spectrometry. Anal. Chem. 2007, 79, 5013–5022. [Google Scholar] [CrossRef]
- Andersson, B.; Holman, R. Pyrrolidides for mass spectrometric determination of the position of the double bond in monounsaturated fatty acids. Lipids 1973, 9, 185–190. [Google Scholar] [CrossRef]
- Capella, P.; Zorzut, C. Determination of double bond position in monounsaturated fatty acid esters by mass spectrometry of their trimethylsilyloxy derivatives. Anal. Chem. 1968, 40, 1458–1463. [Google Scholar] [CrossRef]
- Buser, H.; Arn, H.; Guerin, P.; Rauscher, S. Determination of double bond position in mono-unsaturated acetates by mass spectrometry of dimethyl disulfide adducts. Anal. Chem. 1983, 55, 818–822. [Google Scholar] [CrossRef]
- Kwon, K.; Lee, S.; Oh, D.; Kim, S. Simple determination of double-bond positions in long-chain olefins by cross-metathesis. Angew. Chem. Int. Ed. 2011, 50, 8275–8278. [Google Scholar] [CrossRef]
- Hsu, F.; Turk, J. Elucidation of the double-bond position of long-chain unsaturated fatty acids by multiple-stage linear ion-trap mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2008, 19, 1673–1680. [Google Scholar] [CrossRef]
- Castro-Perez, J.; Roddy, T.; Nibbering, N.; Shah, V.; McLaren, D.; Previs, S.; Attygalle, A.; Herath, K.; Chen, Z.; Wang, S. Localization of fatty acyl and double bond positions in phosphatidylcholines using a dual stage CID fragmentation coupled with ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 2011, 22, 1552–1567. [Google Scholar] [CrossRef]
- Wakelam, M.J.O.; Clark, J. Methods for analyzing phosphoinositides using mass spectrometry. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2011, 1811, 758–762. [Google Scholar] [CrossRef]
- Kilár, A.; Dörnyei, Á.; Kocsis, B. Structural characterization of bacterial lipopolysaccharides with mass spectrometry and on- and off-line separation techniques. Mass Spectrom. Rev. 2013, 32, 90–117. [Google Scholar] [CrossRef]
- Li, F.; Qin, X.; Chen, H.; Qui, L.; Guo, Y.; Liu, H.; Chen, G.; Song, G.; Wang, X.; Li, F. Lipid profiling for early diagnosis and progression of colorectal cancer using direct-infusion electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 24–34. [Google Scholar] [CrossRef]
- Becker, L.; Poutaraud, A.; Hamm, G.; Muller, J.F.; Merdinoglu, D.; Carré, V.; Chaimbault, P. Metabolic study of grapevine leaves infected by downy mildew using negative ion electrospray-Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chim. Acta 2013, 795, 44–51. [Google Scholar]
- Schuhmann, K.; Herzog, R.; Schwudke, D.; Metelmann-Strupat, W.; Bornstein, S.R.; Shevchenko, A. Bottom-up shotgun lipidomics by higher energy collisional dissociation on LTQ Orbitrap mass spectrometers. Anal. Chem. 2011, 83, 5480–5487. [Google Scholar] [CrossRef]
- Bird, S.S.; Marur, V.R.; Sniatynski, M.J.; Greenberg, H.K.; Kristal, B.S. Serum lipidomics profiling using LC-MS and high-energy collisional dissociation fragmentation: Focus on triglyceride detection and characterization. Anal. Chem. 2011, 83, 6648–6657. [Google Scholar] [CrossRef]
- Choi, J.M.; Kim, T.E.; Cho, J.Y.; Lee, H.J.; Jung, B.H. Development of lipidomic platform and phosphatidylcholine retention time index for lipid profiling of rosuvastatin treated human plasma. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 944, 157–165. [Google Scholar]
- Seppanen-Laakso, T.; Oresic, M. How to study lipidomes. J. Mol. Endocrinol. 2009, 42, 185–190. [Google Scholar] [CrossRef]
- Zhao, Z.W.; Xiao, Y.J.; Elson, P.; Tan, H.Y.; Plummer, S.J.; Berk, M.; Aung, P.P.; Lavery, I.C.; Achkar, J.P.; Li, L. Plasma lysophosphatidylcholine levels: Potential biomarkers for colorectal cancer. J. Clin. Oncol. 2007, 25, 2696–2701. [Google Scholar] [CrossRef]
- Poczobutt, J.M.; Gijon, M.; Amin, J.; Hanson, D.; Li, H.; Walker, D.; Weiser-Evans, M.; Lu, X.; Murphy, R.C.; Nemenoff, R.A. Eicosanoid profiling in an orthotopic model of lung cancer progression by mass spectrometry demonstrates selective production of leukotrienes by inflammatory cells of the microenvironment. PLoS One 2013, 8, e79633. [Google Scholar] [CrossRef]
- Han, X.L.; Yang, K.; Gross, R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 2012, 31, 134–178. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, L.; Han, J.; Wang, Z.; Liu, J.; Wei, J.; Xiong, S.; Zhao, Z. Mass Spectrometry Methodology in Lipid Analysis. Int. J. Mol. Sci. 2014, 15, 10492-10507. https://doi.org/10.3390/ijms150610492
Li L, Han J, Wang Z, Liu J, Wei J, Xiong S, Zhao Z. Mass Spectrometry Methodology in Lipid Analysis. International Journal of Molecular Sciences. 2014; 15(6):10492-10507. https://doi.org/10.3390/ijms150610492
Chicago/Turabian StyleLi, Lin, Juanjuan Han, Zhenpeng Wang, Jian'an Liu, Jinchao Wei, Shaoxiang Xiong, and Zhenwen Zhao. 2014. "Mass Spectrometry Methodology in Lipid Analysis" International Journal of Molecular Sciences 15, no. 6: 10492-10507. https://doi.org/10.3390/ijms150610492