Maize ZmRACK1 Is Involved in the Plant Response to Fungal Phytopathogens
Abstract
:1. Introduction
2. Results and Discussion
2.1. Cloning and Bioinformatic Analysis of ZmRACK1
2.2. Expression Pattern of ZmRACK1
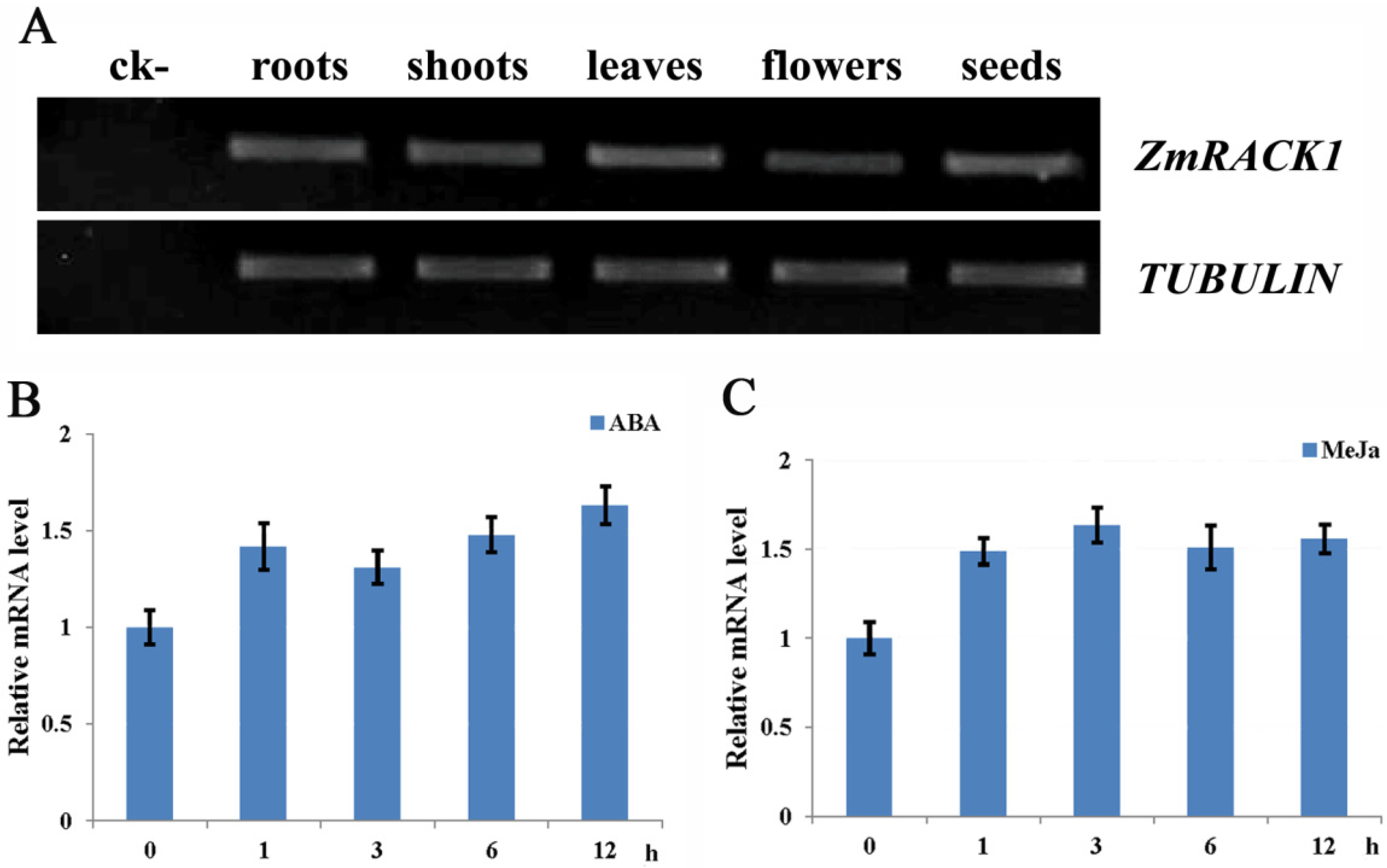
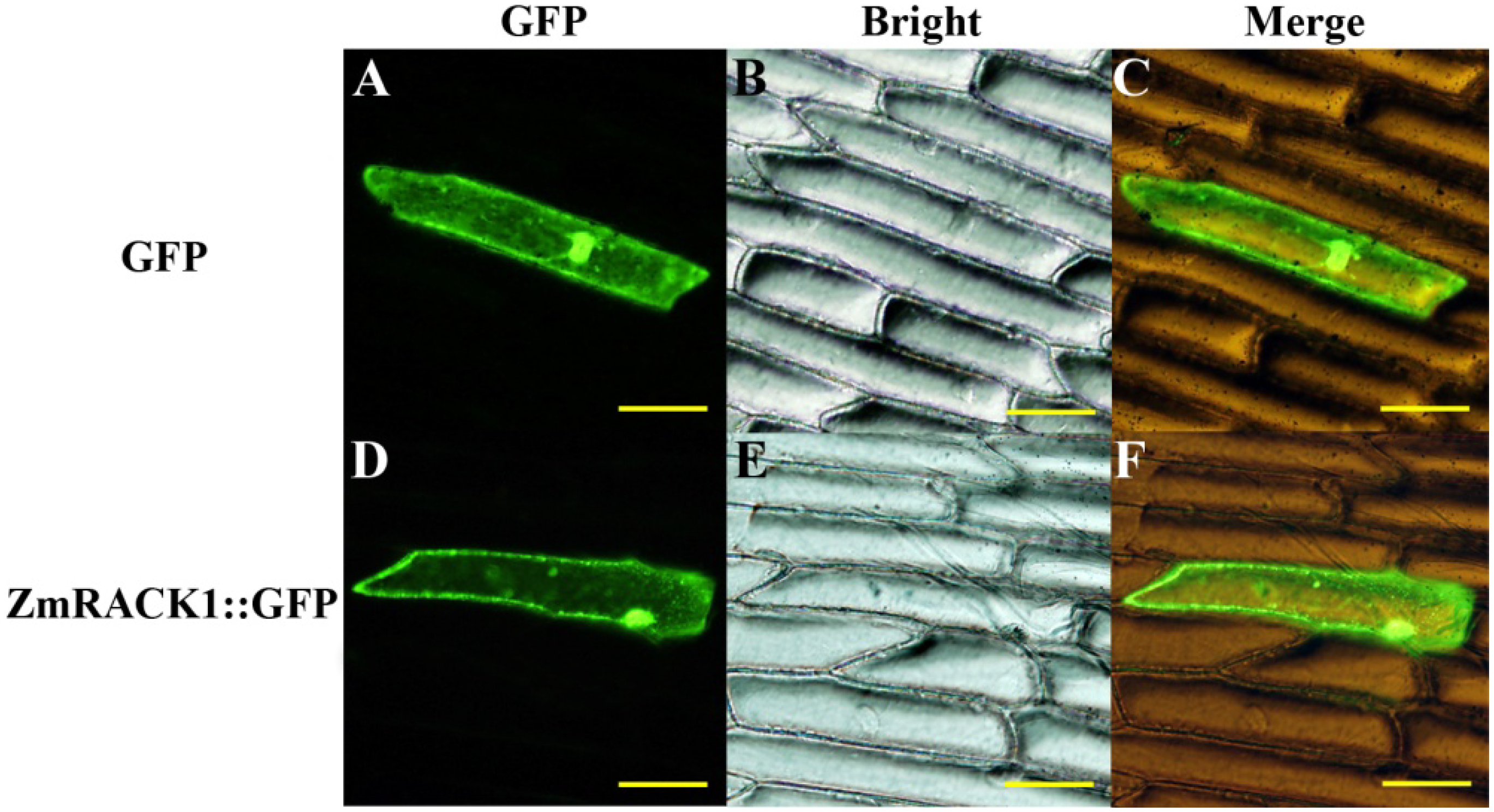
2.3. Intracellular Localization of ZmRACK1
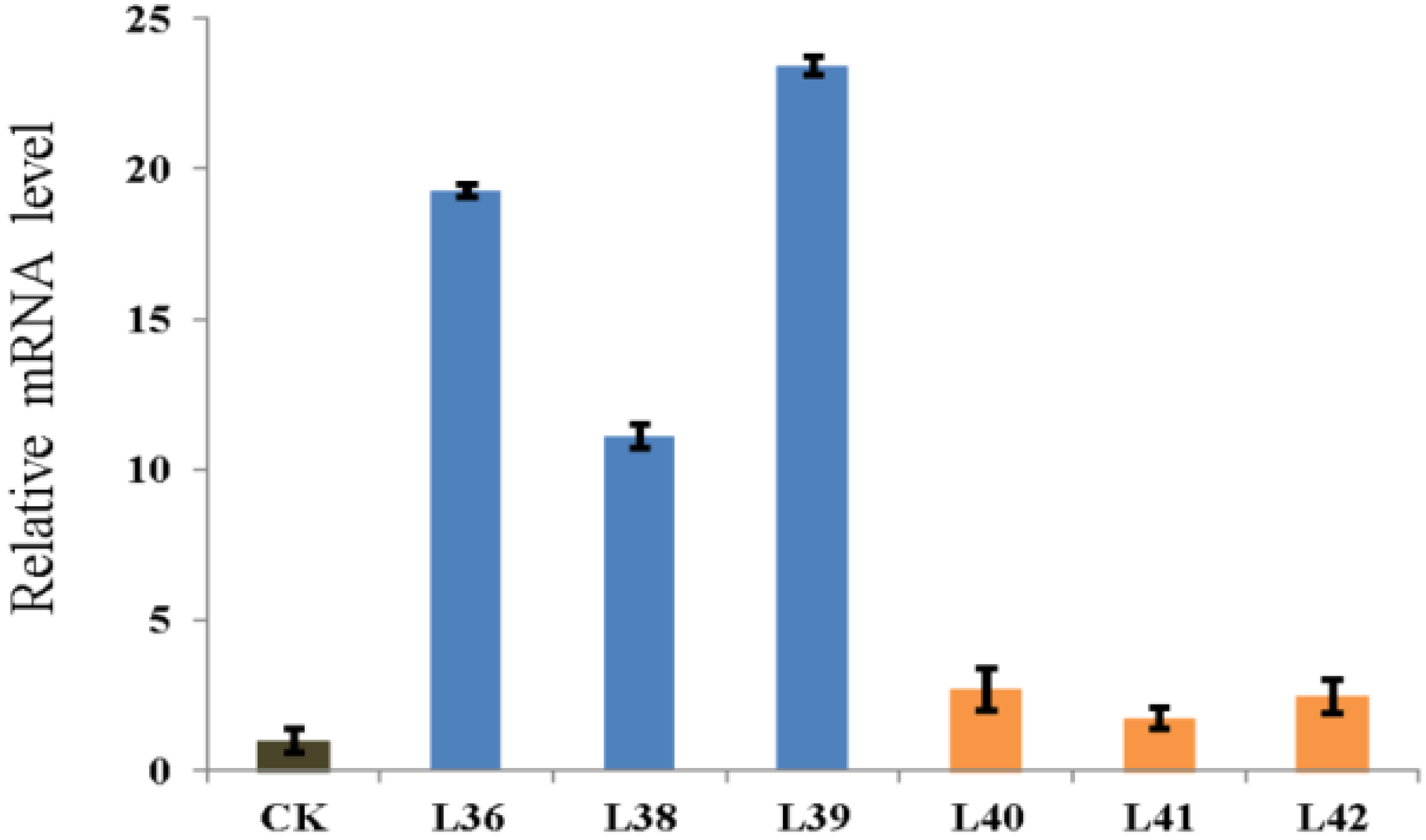
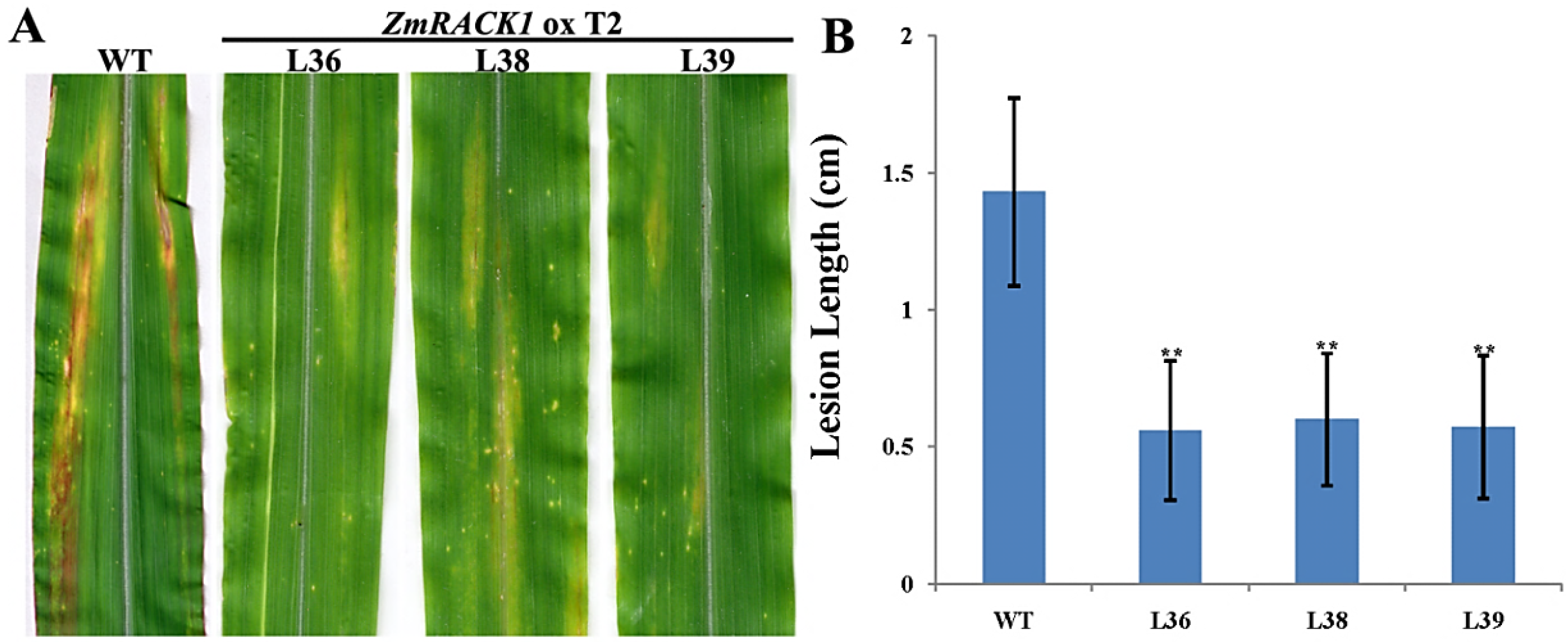
2.4. Overexpression of ZmRACK1 in Maize Confers Resistance to Exserohilum turcicum
| Vector | Line | Plant number of PCR positive | Plant number of PCR negative |
|---|---|---|---|
| pRHU-RACK1 | 36 | 12 | 4 |
| 37 | 12 | 3 | |
| 38 | 14 | 4 | |
| 39 | 11 | 3 | |
| pRHU-RACK1i | 40 | 8 | 7 |
| 41 | 11 | 3 | |
| 42 | 7 | 5 |
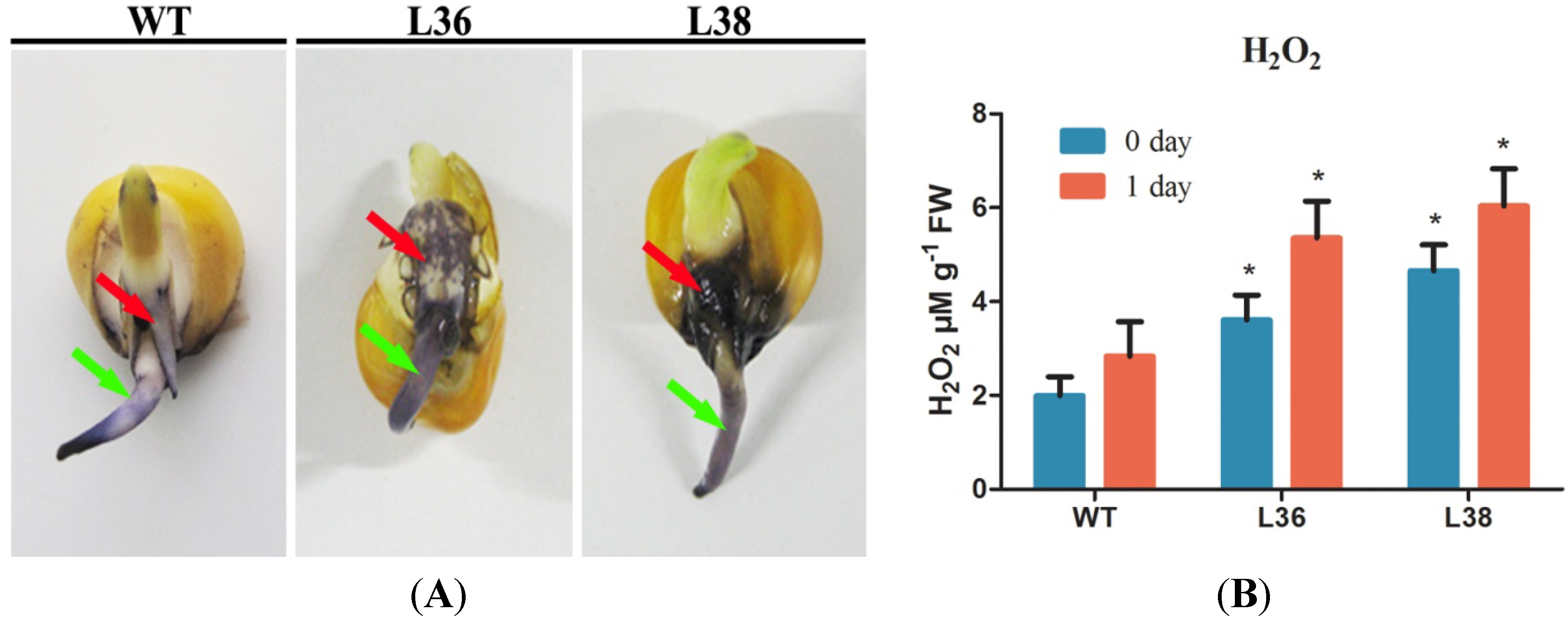
2.5. ZmRACK1 Regulates ROS Production and PR Gene Expression

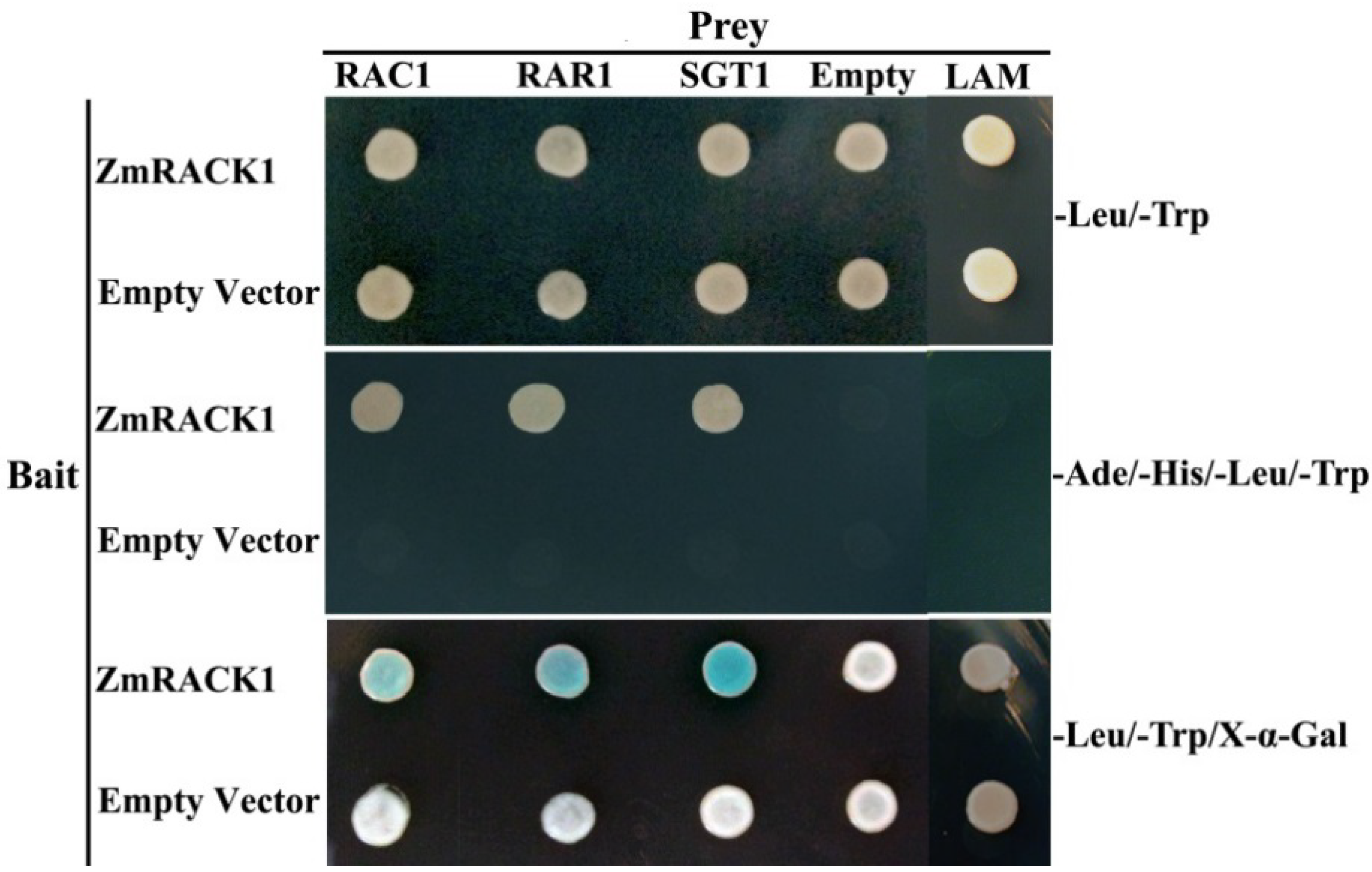
2.6. ZmRACK1 Interacts with RAC1, RAR1 and SGT1
3. Experimental Section
3.1. Plant Materials and Fungal Strain
3.2. Cloning of the ZmRACK1 Gene
3.3. Treatment of Seedlings with MeJA and ABA
3.4. Real-Time PCR
3.5. Transient Expression in Onion Epidermis Cells
3.6. Plasmid Constructs and Maize Transformation
3.7. Infection of Maize Plants with Exserohilum turcicum
3.8. Detection of ROS
3.9. Quantitation of H2O2
3.10. Yeast Two-Hybrid Constructs and Methods
4. Conclusions
Supplementary Files
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Guillemot, F.; Billault, A.; Auffray, C. Physical linkage of a guanine nucleotide-binding protein-related gene to the chicken major histocompatibility complex. Proc. Natl. Acad. Sci. USA 1989, 86, 4594–4598. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; Takahashi, Y.; Nagata, T. Isolation of cDNA of an auxin-regulated gene encoding a G protein β subunit-like protein from tobacco BY-2 cells. Proc. Natl. Acad. Sci. USA 1993, 90, 11152–11156. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Min, J. Structure and function of WD40 domain proteins. Protein Cell 2011, 2, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.G.; Ullah, H.; Temple, B.; Liang, J.; Guo, J.; Alonso, J.M.; Ecker, J.R.; Jones, A.M. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J. Exp. Bot. 2006, 57, 2697–2708. [Google Scholar] [CrossRef] [PubMed]
- Shor, B.; Calaycay, J.; Rushbrook, J.; McLeod, M. Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J. Biol. Chem. 2003, 278, 49119–49128. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.J.; Devkota, B.; Huang, A.D.; Topf, M.; Narayanan, E.; Sali, A.; Harvey, S.C.; Frank, J. Comprehensive molecular structure of the eukaryotic ribosome. Structure 2009, 17, 1591–1604. [Google Scholar] [CrossRef] [PubMed]
- Manuell, A.L.; Yamaguchi, K.; Haynes, P.A.; Milligan, R.A.; Mayfield, S.P. Composition and structure of the 80S ribosome from the green alga Chlamydomonas reinhardtii: 80S Ribosomes are conserved in plants and animals. J. Mol. Biol. 2005, 351, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Link, A.J.; Eng, J.; Schieltz, D.M.; Carmack, E.; Mize, G.J.; Morris, D.R.; Garvik, B.M.; Yates, J.R., III. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 1999, 17, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Chang, I.F.; Szick-Miranda, K.; Pan, S.; Bailey-Serres, J. Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol. 2005, 137, 848–862. [Google Scholar] [CrossRef] [PubMed]
- Mamidipudi, V.; Dhillon, N.K.; Parman, T.; Miller, L.D.; Lee, K.C.; Cartwright, C.A. RACK1 inhibits colonic cell growth by regulating Src activity at cell cycle checkpoints. Oncogene 2007, 26, 2914–2924. [Google Scholar] [CrossRef] [PubMed]
- Fomenkov, A.; Zangen, R.; Huang, Y.P.; Osada, M.; Guo, Z.; Fomenkov, T.; Trink, B.; Sidransky, D.; Ratovitski, E.A. RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle 2004, 3, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Wehner, P.; Shnitsar, I.; Urlaub, H.; Borchers, A. RACK1 is a novel interaction partner of PTK7 that is required for neural tube closure. Development 2011, 138, 1321–1327. [Google Scholar] [CrossRef] [PubMed]
- Kadrmas, J.L.; Smith, M.A.; Pronovost, S.M.; Beckerle, M.C. Characterization of RACK1 function in Drosophila development. Dev. Dyn. 2007, 236, 2207–2215. [Google Scholar]
- Rothberg, K.G.; Burdette, D.L.; Pfannstiel, J.; Jetton, N.; Singh, R.; Ruben, L. The RACK1 homologue from Trypanosoma brucei is required for the onset and progression of cytokinesis. J. Biol. Chem. 2006, 281, 9781–9790. [Google Scholar] [CrossRef] [PubMed]
- McLeod, M.; Shor, B.; Caporaso, A.; Wang, W.; Chen, H.; Hu, L. Cpc2, a fission yeast homologue of mammalian RACK1 protein, interacts with Ran1 (Pat1) kinase to regulate cell cycle progression and meiotic development. Mol. Cell. Biol. 2000, 20, 4016–4027. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Chen, J.G. RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. Biol. Med. Cent. Plant Biol. 2008, 8, 108. [Google Scholar]
- Li, D.H.; Liu, H.; Yang, Y.L.; Zhen, P.P.; Liang, J.S. Down-regulated expression of RACK1 gene by RNA interference enhances drought tolerance in rice. Rice Sci. 2009, 16, 14–20. [Google Scholar] [CrossRef]
- Islas-Flores, T.; Guillen, G.; Islas-Flores, I.; San Roman-Roque, C.; Sanchez, F.; Loza-Tavera, H.; Bearer, E.L.; Villanueva, M.A. Germination behavior, biochemical features and sequence analysis of the RACK1/arcA homolog from Phaseolus vulgaris. Physiol. Plant 2009, 137, 264–280. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.R.; Ron, D.; Kiely, P.A. RACK1, a multifaceted scaffolding protein: Structure and function. Cell Commun. Signal. 2011, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, A.; Chen, L.; Thao, N.P.; Fujiwara, M.; Wong, H.L.; Kuwano, M.; Umemura, K.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 2008, 20, 2265–2279. [Google Scholar] [CrossRef] [PubMed]
- Chou, Y.C.; Chou, C.C.; Chen, Y.K.; Tsai, S.; Hsieh, F.M.; Liu, H.J.; Hseu, T.H. Structure and genomic organization of porcine RACK1 gene. Biochim. Biophys. Acta 1999, 1489, 315–322. [Google Scholar] [CrossRef] [PubMed]
- McKhann, H.I.; Frugier, F.; Petrovics, G.; de la Pena, T.C.; Jurkevitch, E.; Brown, S.; Kondorosi, E.; Kondorosi, A.; Crespi, M. Cloning of a WD-repeat-containing gene from alfalfa (Medicago sativa): A role in hormone-mediated cell division. Plant Mol. Biol. 1997, 34, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, J.; Xi, L.; Huang, W.D.; Liang, J.; Chen, J.G. RACK1 is a negative regulator of ABA responses in Arabidopsis. J. Exp. Bot. 2009, 60, 3819–3833. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, S.; Valerius, O.; Hall, H.; Zeng, Q.; Li, J.F.; Weston, D.J.; Ellis, B.E.; Chen, J.G. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 2011, 155, 370–383. [Google Scholar] [CrossRef] [PubMed]
- He, D.Y.; Vagts, A.J.; Yaka, R.; Ron, D. Ethanol induces gene expression via nuclear compartmentalization of receptor for activated C kinase 1. Mol. Pharmacol. 2002, 62, 272–280. [Google Scholar] [CrossRef] [PubMed]
- He, D.Y.; Neasta, J.; Ron, D. Epigenetic regulation of BDNF expression via the scaffolding protein RACK1. J. Biol. Chem. 2010, 285, 19043–19050. [Google Scholar] [CrossRef] [PubMed]
- Muiru, W.M.K., B.; Tiedemann, A.V.; Mutitu, E.W.; Kimenju, J.W. Race typing and evaluation of aggressiveness of Exserohilum turcicum isolates of Kenyan, German and Austrian origin. World J. Agric. Sci. 2011, 7, 333–340. [Google Scholar]
- Ray, P.D.; Huang, B.W.; Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012, 24, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Adrienne, C.S.; Barbara, J.H. Parallels in fungal pathogenesis on plant and animal hosts: Eukaryot. Cell 2006, 5, 1941–1949. [Google Scholar]
- Niderman, T.; Genetet, I.; Bruyere, T.; Gees, R.; Stintzi, A.; Legrand, M.; Fritig, B.; Mosinger, E. Pathogenesis-related PR-1 proteins are antifungal: Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 1995, 108, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.E.; Fernandes, C.L.; de Souza, O.N.; Salzano, F.M.; Bonatto, S.L.; Freitas, L.B. Molecular modeling of pathogenesis-related proteins of family 5. Cell Biol. Biophys. 2006, 44, 385–394. [Google Scholar]
- Morris, S.W.; Vernooij, B.; Titatarn, S.; Starrett, M.; Thomas, S.; Wiltse, C.C.; Frederiksen, R.A.; Bhandhufalck, A.; Hulbert, S.; Uknes, S. Induced resistance responses in maize. Mol. Plant Microbe Interact. 1998, 11, 643–658. [Google Scholar]
- Kawasaki, T.; Henmi, K.; Ono, E.; Hatakeyama, S.; Iwano, M.; Satoh, H.; Shimamoto, K. The small GTP-binding protein rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 1999, 96, 10922–10926. [Google Scholar] [CrossRef] [PubMed]
- Thao, N.P.; Chen, L.; Nakashima, A.; Hara, S.; Umemura, K.; Takahashi, A.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell 2007, 19, 4035–4045. [Google Scholar] [CrossRef] [PubMed]
- Shirasu, K.; Lahaye, T.; Tan, M.W.; Zhou, F.; Azevedo, C.; Schulze-Lefert, P. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 1999, 99, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Sadanandom, A.; Kitagawa, K.; Freialdenhoven, A.; Shirasu, K.; Schulze-Lefert, P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 2002, 295, 2073–2076. [Google Scholar] [CrossRef] [PubMed]
- Patterson, B.D.; MacRae, E.A.; Ferguson, I.B. Estimation of hydrogen peroxide in plant extracts using titanium(IV). Anal. Biochem. 1984, 139, 487–492. [Google Scholar]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed]
- McCahill, A.; Warwicker, J.; Bolger, G.B.; Houslay, M.D.; Yarwood, S.J. The RACK1 scaffold protein: A dynamic cog in cell response mechanisms. Mol. Pharmacol. 2002, 62, 1261–1273. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.J.; Baillie, G.S.; Yarwood, S.J. Interaction with receptor for activated C-kinase 1 (RACK1) sensitizes the phosphodiesterase PDE4D5 towards hydrolysis of cAMP and activation by protein kinase C. Biochem. J. 2010, 432, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zong, C.S.; Hermanto, U.; Lopez-Bergami, P.; Ronai, Z.; Wang, L.H. RACK1 recruits STAT3 specifically to insulin and insulin-like growth factor 1 receptors for activation, which is important for regulating anchorage-independent growth. Mol. Cell. Biol. 2006, 26, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Ceci, M.; Gaviraghi, C.; Gorrini, C.; Sala, L.A.; Offenhauser, N.; Marchisio, P.C.; Biffo, S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 2003, 426, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Abbasi, F.; Kobori, E.; Fujisawa, Y.; Kato, H.; Iwasaki, Y. Proteomic analysis of rice embryo: An approach for investigating Galpha protein-regulated proteins. Proteomics 2005, 5, 3932–3941. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, B.; Yu, J.; Zhu, D.; Chang, Y.; Zhao, Q. Maize ZmRACK1 Is Involved in the Plant Response to Fungal Phytopathogens. Int. J. Mol. Sci. 2014, 15, 9343-9359. https://doi.org/10.3390/ijms15069343
Wang B, Yu J, Zhu D, Chang Y, Zhao Q. Maize ZmRACK1 Is Involved in the Plant Response to Fungal Phytopathogens. International Journal of Molecular Sciences. 2014; 15(6):9343-9359. https://doi.org/10.3390/ijms15069343
Chicago/Turabian StyleWang, Baosheng, Jingjuan Yu, Dengyun Zhu, Yujie Chang, and Qian Zhao. 2014. "Maize ZmRACK1 Is Involved in the Plant Response to Fungal Phytopathogens" International Journal of Molecular Sciences 15, no. 6: 9343-9359. https://doi.org/10.3390/ijms15069343




