Multiplex Hydrolysis Probe Real-Time PCR for Simultaneous Detection of Hepatitis A Virus and Hepatitis E Virus
Abstract
:1. Introduction
2. Results
2.1. RNA Standard Preparation
2.2. Precision and Reproducibility Analysis
| Template Quantity (Copies/Reaction) | HAV Detection | HEV Detection | ||
|---|---|---|---|---|
| Ct Value (Range) | Average | Ct Value (Range) | Average | |
| 1,000,000 | 17.79–18.01 | 17.89 | 13.56–13.75 | 13.71 |
| 100,000 | 21.11–21.36 | 21.23 | 16.88–17.08 | 16.93 |
| 10,000 | 24.58–24.82 | 24.70 | 20.19–20.45 | 20.33 |
| 1000 | 27.88–28.26 | 28.08 | 23.51–23.92 | 23.76 |
| 100 | 31.22–31.60 | 31.42 | 26.78–27.32 | 27.02 |
| 10 | 34.61–35.11 | 34.83 | 29.77–30.56 | 30.35 |
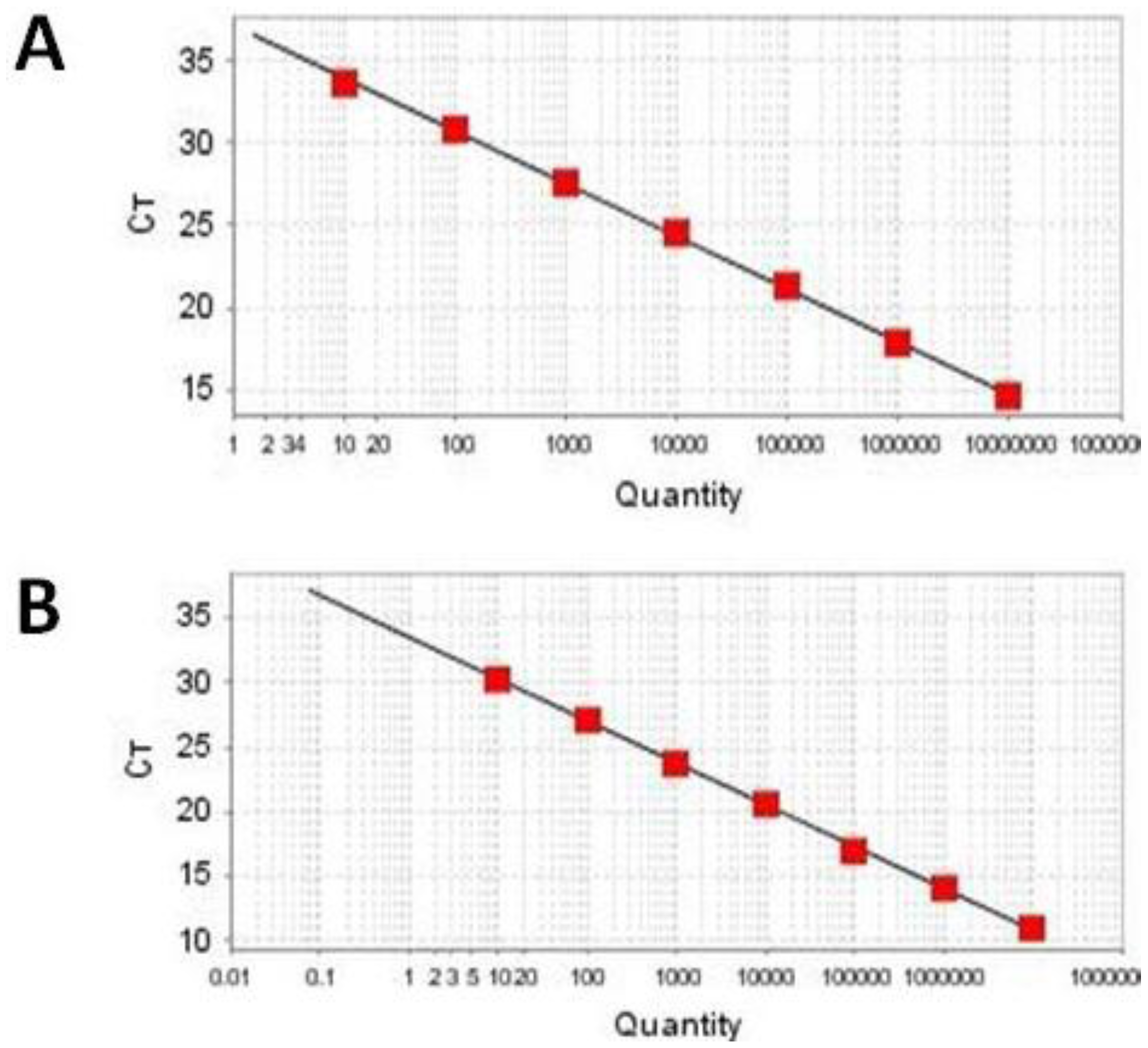
2.3. Sensitivity and Linearity
| Template Quantity (Copies/Reaction) | Ct Value (Average Ct for Triplicate Reactions) | |
|---|---|---|
| HAV Template | HEV Template | |
| 10,000,000 | 14.65 ± 0.09 | 10.28 ± 0.08 |
| 1,000,000 | 17.91±0.14 | 13.71 ± 0.08 |
| 100,000 | 21.29 ± 0.19 | 17.05 ± 0.12 |
| 10,000 | 24.78 ± 0.21 | 20.39 ± 0.18 |
| 1000 | 28.03 ± 0.31 | 23.73 ± 0.31 |
| 100 | 31.39 ± 0.34 | 27.12 ± 0.49 |
| 10 | 34.77 ± 0.52 | 30.54 ± 0.66 |
| 1 | NA | NA |
2.4. Specificity Analysis
2.5. Application of Multiplex Real-Time PCR to Clinical Sera Samples
3. Discussion
4. Methods
4.1. Ethics Issues
4.2. Clinical Specimens from Patients with Acute HAV Infection and HEV Infection
4.3. Viral RNA Extracted from Clinical Sera Specimens
4.4. RNA Standard Preparation
4.5. Primer and Probe Design for HAV-HEV Multiplex Real-Time PCR
| Targeted Viruses | Name | Sequence of Oligonucleotide (5'–3') | Location | Amplicon Length |
|---|---|---|---|---|
| Hepatitis A Virus | Forward primer | GGT AGG CTA CGG GTG AAA C | 393~411 a | 116 nt |
| Reverse primer | CCT CCG GCG TTG AAT GGT TT | 489~508 a | ||
| Probe | FAM-ACA GCG GCG GAT ATT GGT GAG TTG TTA AGA-BHQ | 456~485 a | ||
| Hepatitis E Virus | Forward primer | GGT GGT TTC TGG GGT GAC | 5261~5278 b | 70 nt |
| Reverse primer | AGG GGT TGG TTG GAT GAA | 5313~5330 b | ||
| Probe | Cy5-TGA TTC TCA GCC CTT CGC-BHQ | 5284~5301 b |
4.6. Multiplex Real-Time PCR Conditions
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Mackiewicz, V.; Cammas, A.; Desbois, D.; Marchadier, E.; Pierredon, S.; Beaulieux, F.; Dussaix, E.; Vagner, S.; Roque-Afonso, A.M. Nucleotide variability and translation efficiency of the 5' untranslated region of hepatitis A virus: Update from clinical isolates associated with mild and severe hepatitis. J. Virol. 2010, 84, 10139–10147. [Google Scholar]
- Fujiwara, S.; Yokokawa, Y.; Morino, K.; Hayasaka, K.; Kawabata, M.; Shimizu, T. Chronic hepatitis E: A review of the literature. J. Viral. Hepat. 2014, 21, 78–89. [Google Scholar]
- Sun, Y.; Laird, D.T.; Shieh, Y.C. Temperature-dependent survival of hepatitis A virus during storage of contaminated onions. Appl. Environ. Microbiol. 2012, 78, 4976–4983. [Google Scholar]
- Halliday, M.L.; Kang, L.Y.; Zhou, T.K.; Hu, M.D.; Pan, Q.C.; Fu, T.Y.; Huang, Y.S.; Hu, S.L. An epidemic of hepatitis A attributable to the ingestion of raw clams in Shanghai, China. J. Infect. Dis. 1991, 164, 852–859. [Google Scholar]
- Donnan, E.J.; Fielding, J.E.; Gregory, J.E.; Lalor, K.; Rowe, S.; Goldsmith, P.; Antoniou, M.; Fullerton, K.E.; Knope, K.; Copland, J.G.; et al. multistate outbreak of hepatitis A associated with semidried tomatoes in Australia, 2009. Clin. Infect. Dis. 2012, 54, 775–781. [Google Scholar]
- Arora, D.; Jindal, N.; Shukla, R.K.; Bansal, R. Water borne hepatitis a and hepatitis e in malwa region of punjab, India. J. Clin. Diagn. Res. 2013, 7, 2163–2166. [Google Scholar]
- Walachowski, S.; Dorenlor, V.; Lefevre, J.; Lunazzi, A.; Eono, F.; Merbah, T.; Eveno, E.; Pavio, N.; Rose, N. Risk factors associated with the presence of hepatitis E virus in livers and seroprevalence in slaughter-age pigs: A retrospective study of 90 swine farms in France. Epidemiol. Infect. 2013, 28, 1–11. [Google Scholar]
- Katukiza, A.Y.; Temanu, H.; Chung, J.W.; Foppen, J.W.; Lens, P.N. Genomic copy concentrations of selected waterborne viruses in a slum environment in Kampala, Uganda. J. Water Health 2013, 11, 358–370. [Google Scholar]
- Lee, J.H.; Lee, G.C.; Kim, J.I.; Yi, H.A.; Lee, C.H. Development of a new cell culture-based method and optimized protocol for the detection of enteric viruses. J. Virol. Methods 2013, 191, 16–23. [Google Scholar]
- Jothikumar, N.; Cromeans, T.L.; Sobsey, M.D.; Robertson, B.H. Development and evaluation of a broadly reactive TaqMan assay for rapid detection of hepatitis A virus. Appl. Environ. Microbiol. 2005, 71, 3359–3363. [Google Scholar]
- Costafreda, M.I.; Bosch, A.; Pinto, R.M. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 2006, 72, 3846–3855. [Google Scholar]
- Mokhtari, C.; Marchadier, E.; Haim-Boukobza, S.; Jeblaoui, A.; Tesse, S.; Savary, J.; Roque-Afonso, A.M. Comparison of real-time RT-PCR assays for hepatitis E virus RNA detection. J. Clin. Virol. 2013, 58, 36–40. [Google Scholar]
- Traore, K.A.; Rouamba, H.; Nebie, Y.; Sanou, M.; Traore, A.S.; Barro, N.; Roques, P. Seroprevalence of fecal-oral transmitted hepatitis A and E virus antibodies in Burkina Faso. PLoS One 2012, 7, e48125. [Google Scholar]
- Orru, G.; Masia, G.; Romano, L.; Piras, V.; Coppola, R.C. Detection and quantitation of hepatitis E virus in human faeces by real-time quantitative PCR. J. Virol. Methods 2004, 118, 77–82. [Google Scholar]
- Jothikumar, N.; Cromeans, T.L.; Robertson, B.H.; Meng, X.J.; Hill, V.R. A broadly reactive one-step real-time RT-PCR assay for rapid and sensitive detection of hepatitis E virus. J. Virol. Methods 2006, 131, 65–71. [Google Scholar]
- Irshad, M.; Ansari, M.A.; Irshad, K.; Lingaiah, R. Novel single-step multiplex real-time polymerase chain reaction assay for simultaneous quantification of hepatitis virus A, B, C, and E in serum. J. Gastroenterol. Hepatol. 2013, 28, 1869–1876. [Google Scholar]
- Qiu, F.; Zheng, H.H.; Yi, Y.; Jia, Z.Y.; Cao, J.Y.; Bi, S.L. Comparative evaluation of a novel TaqMan real-time reverse transcription-polymerase chain reaction assay for hepatitis A virus detection. J. Int. Med. Res. 2013, 41, 427–434. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Qiu, F.; Cao, J.; Su, Q.; Yi, Y.; Bi, S. Multiplex Hydrolysis Probe Real-Time PCR for Simultaneous Detection of Hepatitis A Virus and Hepatitis E Virus. Int. J. Mol. Sci. 2014, 15, 9780-9788. https://doi.org/10.3390/ijms15069780
Qiu F, Cao J, Su Q, Yi Y, Bi S. Multiplex Hydrolysis Probe Real-Time PCR for Simultaneous Detection of Hepatitis A Virus and Hepatitis E Virus. International Journal of Molecular Sciences. 2014; 15(6):9780-9788. https://doi.org/10.3390/ijms15069780
Chicago/Turabian StyleQiu, Feng, Jingyuan Cao, Qiudong Su, Yao Yi, and Shengli Bi. 2014. "Multiplex Hydrolysis Probe Real-Time PCR for Simultaneous Detection of Hepatitis A Virus and Hepatitis E Virus" International Journal of Molecular Sciences 15, no. 6: 9780-9788. https://doi.org/10.3390/ijms15069780




