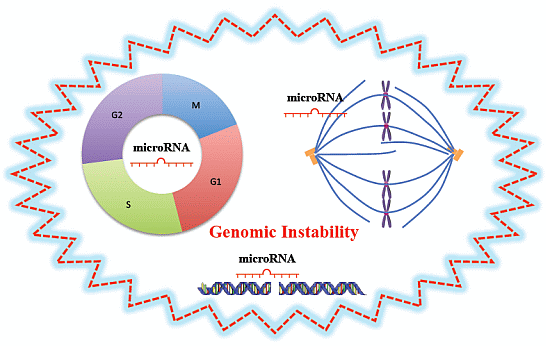
MicroRNAs, Genomic Instability and Cancer
Abstract
:1. Introduction
| miRNAs | Cancer Type | Targets | Function | Category | Effect | Reference |
|---|---|---|---|---|---|---|
| miR-16 | Breast cancer | WIP1 | Enhances response to DNA damage | DNA damage response | Prevents genomic instability | [12] |
| miR-24 | Hematopoietic malignancies | E2F2, CDK1, CDK4 | Increases cell population in the G1 phase | Cell cycle | Enhances genomic instability | [13] |
| miR-24 | Hematopoietic malignancies | H2AX | Impairs DNA repair | DNA damage response | Enhances genomic instability | [14] |
| miR-24 | Hepatocellular carcinoma | AURKB | Impairs chromosome separation | Mitotic events | Enhances genomic instability | [15] |
| miR-29 | Breast cancer, colon cancer | PIK3R1 and CDC42 | Increases p53 stability | DNA damage response | Prevents genomic instability | [16] |
| miR-34 | Colon cancer, hepatocellular carcinoma | CCND1, CCNE2, CDK4, MET, MYC, and SIRT1 | Promotes DNA damage repair, or induces apoptosis | DNA damage response | Prevents genomic instability | [17,18,19,20] |
| miR-96 | Osteosarcoma, breast cancer | RAD51 | Impairs HR repair | DNA damage response | Enhances genomic instability | [21] |
| miR-100 | Nasopharyngeal cancer | PLK1 | Loss of miR-100 promotes mitotic catastrophe | Mitotic events | Prevents genomic instability | [22] |
| miR-100 | Glioblastoma | ATM | Impairs response to DNA damage | DNA damage response | Enhances genomic instability | [23] |
| miR-101 | Glioblastoma | ATM | Impairs response to DNA damage | DNA damage response | Enhances genomic instability | [24] |
| miR-101 | Glioblastoma | PRKDC | Impairs NHEJ repair | DNA damage response | Enhances genomic instability | [24] |
| miR-103 | Hepatocellular carcinoma, osteosarcoma, ovarian cancer | RAD51 | Impairs HR repair | DNA damage response | Enhances genomic instability | [25] |
| miR-106b-25 cluster | Colon cancer | p21 | Promotes the G1-to-S transition | Cell cycle | Enhances genomic instability | [26] |
| miR-107 | Hepatocellular carcinoma, osteosarcoma, ovarian cancer | RAD51 | Impairs HR repair | DNA damage response | Enhances genomic instability | [25] |
| miR-122 | Hepatocellular carcinoma | CCND1 | Upregualtes p53 expression | DNA damage response | Prevents genomic instability | [27] |
| miR-125b | Head and neck cancer | MXD1 | Delays mitotic progression at metaphase | Mitotic events | Enhances genomic instability | [28] |
| miR-125b | Neuroblastoma | P53 | Reduces p53 expression | DNA damage response | Enhances genomic instability | [29] |
| miR-138 | Osteosarcoma | H2AX | Impairs response to DNA damage | DNA damage response | Enhances genomic instability | [30] |
| miR-148b | Breast cancer | RAD51 | Impairs HR repair | DNA damage response | Enhances genomic instability | [31] |
| miR-155 | Colon cancer, breast cancer | MLH1, MSH2, MSH6, TERF1 | Promotes microsatellite instability; impairs telomere integrity | DNA damage response | Enhances genomic instability | [32,33] |
| miR-182 | Colon cancer, breast cancer | BRCA1 | Impairs response to DNA damage | DNA damage response | Enhances genomic instability | [34] |
| miR-193b | Breast cancer | RAD51 | Impairs HR repair | DNA damage response | Enhances genomic instability | [31] |
| miR-193b | Breast cancer | BRCA1 and BRCA2 | Impairs HR repair | DNA damage response | Enhances genomic instability | [31] |
| miR-210 | Breast cancer, hepatocellular carcinoma | RAD52 | Impairs HR repair | DNA damage response | Enhances genomic instability | [35] |
| miR-221/222 | Thyroid papillary carcinoma, glioblastoma | p27 | Increases cell population in the S phase | Cell cycle | Enhances genomic instability | [36,37] |
| miR-372 | Testicular germ cell tumor, cervical cancer | LATS2, CDK2, CCNA1 | Compromises p53-mediated CDK inhibition; Delays entrance into G2/M from S phase | Cell cycle | Enhances genomic instability | [38,39] |
| miR-373 | Breast cancer, hepatocellular carcinoma | RAD52 and RAD23B | Impairs HR repair | DNA damage response | Enhances genomic instability | [35] |
| miR-421 | Neuroblastoma | ATM | Impairs response to DNA damage | DNA damage response | Enhances genomic instability | [40] |
| miR-504 | Colon cancer | P53 | Reduces p53 expression | DNA damage response | Enhances genomic instability | [41] |
| miR-1255b | Breast cancer | BRCA1 and BRCA2 | Impairs HR repair | DNA damage response | Enhances genomic instability | [31] |
2. MiRNAs Are Frequently Located in the Fragile Sites of the Genome
3. MiRNAs in Genomic Instability and Cancer
3.1. MiRNAs and Cell Cycle
3.1.1. MiR-372/LATS2, CDK2, CCNA1
3.1.2. MiR-106b-25 Cluster/p21
3.1.3. MiR-221/222/p27
3.1.4. MiR-24/E2F2, CDK1, CDK4
3.2. MiRNAs and DNA Damage Response
3.2.1. MiR-155/MLH1, MSH2, MSH6, TERF1
3.2.2. MiR-29/34/122/125b/504/p53 and p53-Regualted Genes
3.2.3. MiR-182/193b/1255b/BRCA1
3.2.4. MiR-24/138/H2AX
3.2.5. miR-16/100/101/421/ATM
3.2.6. MiR-96/103/107/148b/193b/RAD51
3.2.7. MiR-210/373/RAD52
3.3. MiRNAs and Mitotic Events
3.3.1. MiR-125b/MXD1
3.3.2. MiR-100/PLK1
3.3.3. MiR-24/AURKB
4. Conclusion Notes
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bernstein, B.E.; Birney, E.; Dunham, I.; Green, E.D.; Gunter, C.; Snyder, M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. MiR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Ling, H.; Zhang, W.; Calin, G.A. Principles of microRNA involvement in human cancers. Chin. J. Cancer 2011, 30, 739–748. [Google Scholar] [CrossRef]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef]
- Pichler, M.; Ress, A.L.; Winter, E.; Stiegelbauer, V.; Karbiener, M.; Schwarzenbacher, D.; Scheideler, M.; Ivan, C.; Jahn, S.W.; Kiesslich, T.; et al. MiR-200a regulates epithelial to mesenchymal transition-related gene expression and determines prognosis in colorectal cancer patients. Br. J. Cancer 2014, 110, 1614–1621. [Google Scholar] [CrossRef]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef]
- Zhang, X.; Wan, G.; Mlotshwa, S.; Vance, V.; Berger, F.G.; Chen, H.; Lu, X. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010, 70, 7176–7186. [Google Scholar] [CrossRef]
- Lal, A.; Navarro, F.; Maher, C.A.; Maliszewski, L.E.; Yan, N.; O’Day, E.; Chowdhury, D.; Dykxhoorn, D.M.; Tsai, P.; Hofmann, O.; et al. MiR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3'-UTR microRNA recognition elements. Mol. Cell 2009, 35, 610–625. [Google Scholar] [CrossRef]
- Lal, A.; Pan, Y.; Navarro, F.; Dykxhoorn, D.M.; Moreau, L.; Meire, E.; Bentwich, Z.; Lieberman, J.; Chowdhury, D. MiR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 2009, 16, 492–498. [Google Scholar] [CrossRef]
- Akbari Moqadam, F.; Boer, J.M.; Lange-Turenhout, E.A.; Pieters, R.; den Boer, M.L. Altered expression of miR-24, miR-126 and miR-365 does not affect viability of childhood TCF3-rearranged leukemia cells. Leukemia 2014, 28, 1008–1014. [Google Scholar] [CrossRef]
- Park, S.Y.; Lee, J.H.; Ha, M.; Nam, J.W.; Kim, V.N. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat. Struct. Mol. Biol. 2009, 16, 23–29. [Google Scholar]
- He, L.; He, X.; Lim, L.P.; de Stanchina, E.; Xuan, Z.; Liang, Y.; Xue, W.; Zender, L.; Magnus, J.; Ridzon, D.; et al. A microRNA component of the p53 tumour suppressor network. Nature 2007, 447, 1130–1134. [Google Scholar] [CrossRef]
- Hermeking, H. MicroRNAs in the p53 network: Micromanagement of tumour suppression. Nat. Rev. Cancer 2012, 12, 613–626. [Google Scholar] [CrossRef]
- Chang, T.C.; Wentzel, E.A.; Kent, O.A.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 2007, 26, 745–752. [Google Scholar] [CrossRef]
- Kato, M.; Paranjape, T.; Muller, R.U.; Nallur, S.; Gillespie, E.; Keane, K.; Esquela-Kerscher, A.; Weidhaas, J.B.; Slack, F.J. The miR-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 2009, 28, 2419–2424. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.W.; Calses, P.; Kemp, C.J.; Taniguchi, T. MiR-96 downregulates REV1 and RAD51 to promote cellular sensitivity to cisplatin and PARP inhibition. Cancer Res. 2012, 72, 4037–4046. [Google Scholar] [CrossRef]
- Shi, W.; Alajez, N.M.; Bastianutto, C.; Hui, A.B.; Mocanu, J.D.; Ito, E.; Busson, P.; Lo, K.W.; Ng, R.; Waldron, J.; et al. Significance of Plk1 regulation by miR-100 in human nasopharyngeal cancer. Int. J. Cancer 2010, 126, 2036–2048. [Google Scholar]
- Ng, W.L.; Yan, D.; Zhang, X.; Mo, Y.Y.; Wang, Y. Over-expression of miR-100 is responsible for the low-expression of ATM in the human glioma cell line: M059J. DNA Repair 2010, 9, 1170–1175. [Google Scholar] [CrossRef]
- Yan, D.; Ng, W.L.; Zhang, X.; Wang, P.; Zhang, Z.; Mo, Y.Y.; Mao, H.; Hao, C.; Olson, J.J.; Curran, W.J.; et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One 2010, 5, e11397. [Google Scholar] [CrossRef]
- Huang, J.W.; Wang, Y.; Dhillon, K.K.; Calses, P.; Villegas, E.; Mitchell, P.S.; Tewari, M.; Kemp, C.J.; Taniguchi, T. Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity. Mol. Cancer Res. 2013, 11, 1564–1573. [Google Scholar] [CrossRef]
- Ivanovska, I.; Ball, A.S.; Diaz, R.L.; Magnus, J.F.; Kibukawa, M.; Schelter, J.M.; Kobayashi, S.V.; Lim, L.; Burchard, J.; Jackson, A.L.; et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 2008, 28, 2167–2174. [Google Scholar] [CrossRef]
- Fornari, F.; Gramantieri, L.; Giovannini, C.; Veronese, A.; Ferracin, M.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Croce, C.M.; Tavolari, S.; et al. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009, 69, 5761–5767. [Google Scholar] [CrossRef]
- Bhattacharjya, S.; Nath, S.; Ghose, J.; Maiti, G.P.; Biswas, N.; Bandyopadhyay, S.; Panda, C.K.; Bhattacharyya, N.P.; Roychoudhury, S. miR-125b promotes cell death by targeting spindle assembly checkpoint gene MAD1 and modulating mitotic progression. Cell Death Differ. 2013, 20, 430–442. [Google Scholar] [CrossRef]
- Le, M.T.; Teh, C.; Shyh-Chang, N.; Xie, H.; Zhou, B.; Korzh, V.; Lodish, H.F.; Lim, B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009, 23, 862–876. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.W.; Li, M.; Cavenee, W.K.; Mitchell, P.S.; Zhou, X.; Tewari, M.; Furnari, F.B.; Taniguchi, T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol. Cancer Res. 2011, 9, 1100–1111. [Google Scholar] [CrossRef]
- Choi, Y.E.; Pan, Y.; Park, E.; Konstantinopoulos, P.; De, S.; D’Andrea, A.; Chowdhury, D. MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. eLife 2014, 3, e02445. [Google Scholar]
- Valeri, N.; Gasparini, P.; Fabbri, M.; Braconi, C.; Veronese, A.; Lovat, F.; Adair, B.; Vannini, I.; Fanini, F.; Bottoni, A.; et al. Modulation of mismatch repair and genomic stability by miR-155. Proc. Natl. Acad. Sci. USA 2010, 107, 6982–6987. [Google Scholar] [CrossRef]
- Dinami, R.; Ercolani, C.; Petti, E.; Piazza, S.; Ciani, Y.; Sestito, R.; Sacconi, A.; Biagioni, F.; le Sage, C.; Agami, R.; et al. miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res. 2014, 74, 4145–4156. [Google Scholar] [CrossRef]
- Moskwa, P.; Buffa, F.M.; Pan, Y.; Panchakshari, R.; Gottipati, P.; Muschel, R.J.; Beech, J.; Kulshrestha, R.; Abdelmohsen, K.; Weinstock, D.M.; et al. MiR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 2011, 41, 210–220. [Google Scholar] [CrossRef]
- Crosby, M.E.; Kulshreshtha, R.; Ivan, M.; Glazer, P.M. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009, 69, 1221–1229. [Google Scholar] [CrossRef]
- Visone, R.; Russo, L.; Pallante, P.; de Martino, I.; Ferraro, A.; Leone, V.; Borbone, E.; Petrocca, F.; Alder, H.; Croce, C.M.; et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr.-Relat. Cancer 2007, 14, 791–798. [Google Scholar] [CrossRef]
- Gillies, J.K.; Lorimer, I.A. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle 2007, 6, 2005–2009. [Google Scholar] [CrossRef]
- Voorhoeve, P.M.; le Sage, C.; Schrier, M.; Gillis, A.J.; Stoop, H.; Nagel, R.; Liu, Y.P.; van Duijse, J.; Drost, J.; Griekspoor, A.; et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006, 124, 1169–1181. [Google Scholar] [CrossRef]
- Tian, R.Q.; Wang, X.H.; Hou, L.J.; Jia, W.H.; Yang, Q.; Li, Y.X.; Liu, M.; Li, X.; Tang, H. MicroRNA-372 is down-regulated and targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human cervical cancer, which may contribute to tumorigenesis. J. Biol. Chem. 2011, 286, 25556–25563. [Google Scholar]
- Hu, H.; Du, L.; Nagabayashi, G.; Seeger, R.C.; Gatti, R.A. ATM is down-regulated by N-Myc-regulated microRNA-421. Proc. Natl. Acad. Sci. USA 2010, 107, 1506–1511. [Google Scholar] [CrossRef]
- Hu, W.; Chan, C.S.; Wu, R.; Zhang, C.; Sun, Y.; Song, J.S.; Tang, L.H.; Levine, A.J.; Feng, Z. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell 2010, 38, 689–699. [Google Scholar] [CrossRef]
- Landau, D.A.; Slack, F.J. MicroRNAs in mutagenesis, genomic instability, and DNA repair. Semin. Oncol. 2011, 38, 743–751. [Google Scholar] [CrossRef]
- Wang, Y.; Taniguchi, T. MicroRNAs and DNA damage response: Implications for cancer therapy. Cell Cycle 2013, 12, 32–42. [Google Scholar] [CrossRef]
- Chowdhury, D.; Choi, Y.E.; Brault, M.E. Charity begins at home: Non-coding RNA functions in DNA repair. Nat. Rev. Mol. Cell Biol. 2013, 14, 181–189. [Google Scholar] [CrossRef]
- Tessitore, A.; Cicciarelli, G.; del Vecchio, F.; Gaggiano, A.; Verzella, D.; Fischietti, M.; Vecchiotti, D.; Capece, D.; Zazzeroni, F.; Alesse, E. MicroRNAs in the DNA damage/repair network and cancer. Int. J. Genomics 2014, 2014, 820248. [Google Scholar]
- Wouters, M.D.; van Gent, D.C.; Hoeijmakers, J.H.; Pothof, J. MicroRNAs, the DNA damage response and cancer. Mutat. Res. 2011, 717, 54–66. [Google Scholar] [CrossRef]
- Yunis, J.J.; Soreng, A.L. Constitutive fragile sites and cancer. Science 1984, 226, 1199–1204. [Google Scholar]
- Arlt, M.F.; Casper, A.M.; Glover, T.W. Common fragile sites. Cytogenet. Genome Res. 2003, 100, 92–100. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef]
- Jimenez-Wences, H.; Peralta-Zaragoza, O.; Fernandez-Tilapa, G. Human papilloma virus, DNA methylation and microRNA expression in cervical cancer (Review). Oncol. Rep. 2014, 31, 2467–2476. [Google Scholar]
- Wang, C.L.; Wang, B.B.; Bartha, G.; Li, L.; Channa, N.; Klinger, M.; Killeen, N.; Wabl, M. Activation of an oncogenic microRNA cistron by provirus integration. Proc. Natl. Acad. Sci. USA 2006, 103, 18680–18684. [Google Scholar]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Diaz-Beya, M.; Navarro, A.; Ferrer, G.; Diaz, T.; Gel, B.; Camos, M.; Pratcorona, M.; Torrebadell, M.; Rozman, M.; Colomer, D.; et al. Acute myeloid leukemia with translocation (8;16) (p11;p13) and MYST3-CREBBP rearrangement harbors a distinctive microRNA signature targeting RET proto-oncogene. Leukemia 2013, 27, 595–603. [Google Scholar] [CrossRef]
- Robbiani, D.F.; Bunting, S.; Feldhahn, N.; Bothmer, A.; Camps, J.; Deroubaix, S.; McBride, K.M.; Klein, I.A.; Stone, G.; Eisenreich, T.R.; et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol. Cell 2009, 36, 631–641. [Google Scholar] [CrossRef]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Aylon, Y.; Michael, D.; Shmueli, A.; Yabuta, N.; Nojima, H.; Oren, M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006, 20, 2687–2700. [Google Scholar] [CrossRef]
- Polyak, K.; Lee, M.H.; Erdjument-Bromage, H.; Koff, A.; Roberts, J.M.; Tempst, P.; Massague, J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 1994, 78, 59–66. [Google Scholar] [CrossRef]
- Costinean, S.; Zanesi, N.; Pekarsky, Y.; Tili, E.; Volinia, S.; Heerema, N.; Croce, C.M. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA 2006, 103, 7024–7029. [Google Scholar]
- Malkin, D.; Li, F.P.; Strong, L.C.; Fraumeni, J.F., Jr.; Nelson, C.E.; Kim, D.H.; Kassel, J.; Gryka, M.A.; Bischoff, F.Z.; Tainsky, M.A.; et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 1990, 250, 1233–1238. [Google Scholar]
- Deng, C.X. BRCA1: Cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006, 34, 1416–1426. [Google Scholar] [CrossRef]
- Zhang, X.; Wan, G.; Berger, F.G.; He, X.; Lu, X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol. Cell 2011, 41, 371–383. [Google Scholar] [CrossRef]
- Neijenhuis, S.; Bajrami, I.; Miller, R.; Lord, C.J.; Ashworth, A. Identification of miRNA modulators to PARP inhibitor response. DNA Repair 2013, 12, 394–402. [Google Scholar] [CrossRef]
- Vitale, I.; Galluzzi, L.; Castedo, M.; Kroemer, G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 2011, 12, 385–392. [Google Scholar] [CrossRef]
- Strebhardt, K.; Ullrich, A. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev.Cancer 2006, 6, 321–330. [Google Scholar] [CrossRef]
- Li, Y.; Xu, F.L.; Lu, J.; Saunders, W.S.; Prochownik, E.V. Widespread genomic instability mediated by a pathway involving glycoprotein Ib alpha and Aurora B kinase. J. Biol. Chem. 2010, 285, 13183–13192. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Vincent, K.; Pichler, M.; Lee, G.-W.; Ling, H. MicroRNAs, Genomic Instability and Cancer. Int. J. Mol. Sci. 2014, 15, 14475-14491. https://doi.org/10.3390/ijms150814475
Vincent K, Pichler M, Lee G-W, Ling H. MicroRNAs, Genomic Instability and Cancer. International Journal of Molecular Sciences. 2014; 15(8):14475-14491. https://doi.org/10.3390/ijms150814475
Chicago/Turabian StyleVincent, Kimberly, Martin Pichler, Gyeong-Won Lee, and Hui Ling. 2014. "MicroRNAs, Genomic Instability and Cancer" International Journal of Molecular Sciences 15, no. 8: 14475-14491. https://doi.org/10.3390/ijms150814475