Sperm and Spermatids Contain Different Proteins and Bind Distinct Egg Factors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sperm and Spermatids Differ in Their Nuclear Protein Composition
2.1.1. Sperm and Spermatids Contain Different Proteins


2.1.2. Sperm-Specific Proteins Contain Several Factors Which May Support Subsequent Embryonic Development
2.1.3. Chromatin and Nuclear Proteins Are Enriched in Sperm
2.2. Sperm and Spermatids Attract Distinct Egg Factors
2.2.1. Dramatic Changes in Protein Composition after the Egg Extract Treatment


2.2.2. Many Egg Proteins Bind Differentially to Sperm and to Spermatid Chromatin

2.2.3. Known Chromatin Remodelling Factors and Replication Proteins Were Incorporated from Egg Extracts into both Sperm and Spermatids
2.2.4. Distinct Protein Isoforms Identified in Extract-Treated Sperm- and Spermatids
2.2.5. Many Egg-Derived Chromatin Factors Bind Preferentially to Sperm Nuclei
2.2.6. GO (Gene Ontology) Analysis Confirms the Overrepresentation of Egg Chromatin Remodelling Factors Binding Specifically to Sperm Nuclei
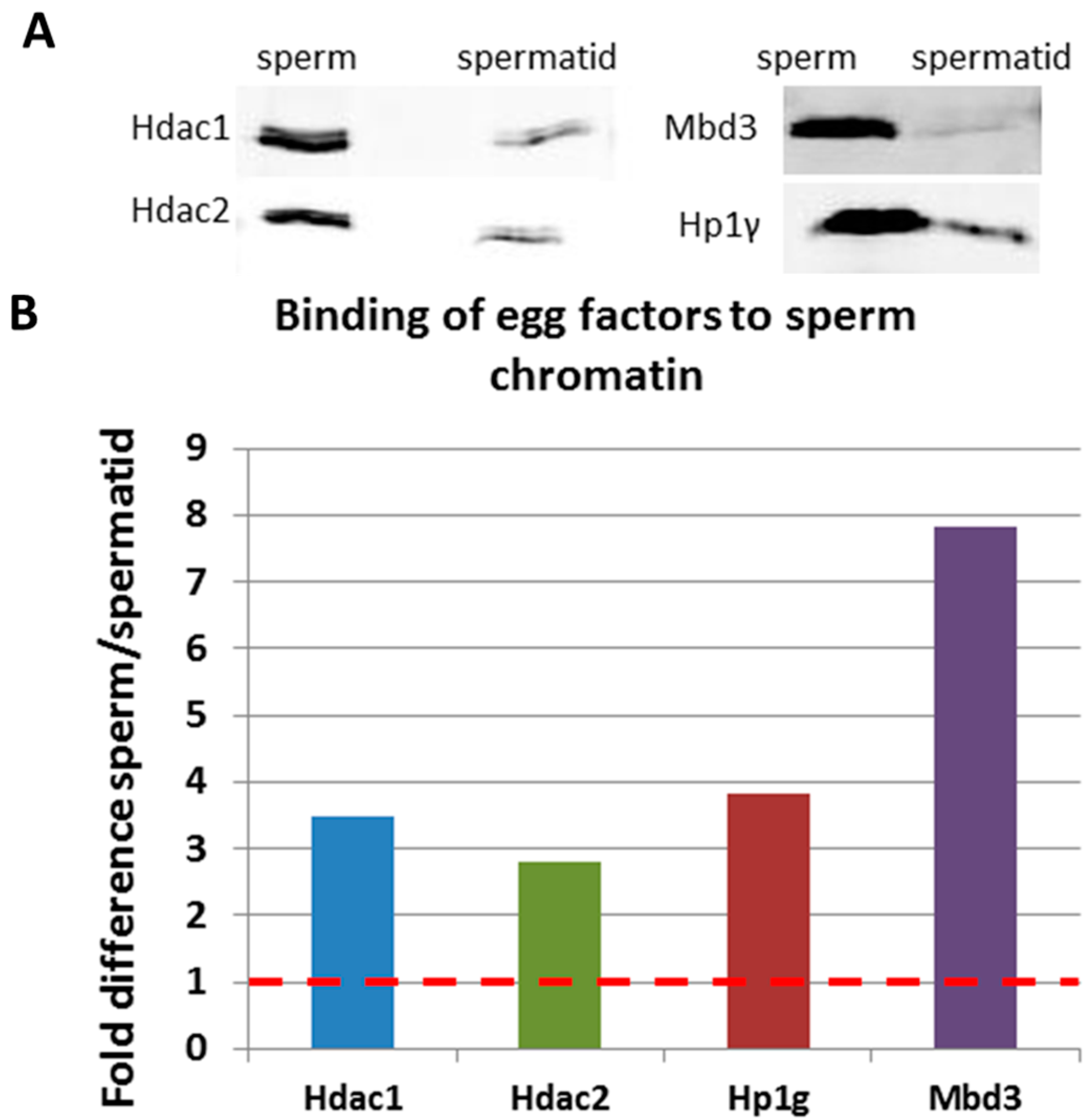
2.3. Validation of Mass Spectrometry Results by Immunoblotting
3. Experimental Section
3.1. Isolation and Permeabilization of Sperm and Spermatids
3.2. Isolation of Proteins and Sample Preparation for 2D-DIGE
3.3. 2D-DIGE Analysis
3.4. Spot Excision and Protein Preparation for Mass Spectrometry Analysis
3.5. LC-MS/MS Analysis and Protein Identification
3.6. Interphase Egg Extract Preparation
3.7. Egg Extract Treatment and Protein Isolation for Mass Spectrometry Analysis of Egg Proteins Incorporated into Sperm or Spermatid Chromatin
3.8. Immunoblotting Analysis
4. Conclusions
Supplementary Materials
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- De Rooij, D.G. Proliferation and differentiation of spermatogonial stem cells. Reproduction 2001, 121, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, J.; Reynoird, N.; Montellier, E.; Boussouar, F.; Rousseaux, S.; Khochbin, S. From meiosis to postmeiotic events: The secrets of histone disappearance. FEBS J. 2010, 277, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Shen, J.; Xia, Z.; Zhang, R.; Zhang, P.; Zhao, C.; Xing, J.; Chen, L.; Chen, W.; Lin, M.; et al. Proteomic analysis of proteins involved in spermiogenesis in mouse. J. Proteome Res. 2010, 9, 1246–1256. [Google Scholar] [CrossRef] [PubMed]
- De Mateo, S.; Castillo, J.; Estanyol, J.M.; Ballesca, J.L.; Oliva, R. Proteomic characterization of the human sperm nucleus. Proteomics 2011, 11, 2714–2726. [Google Scholar] [CrossRef] [PubMed]
- Oliva, R.; Castillo, J. Proteomics and the genetics of sperm chromatin condensation. Asian J. Androl. 2011, 13, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Govin, J.; Gaucher, J.; Ferro, M.; Debernardi, A.; Garin, J.; Khochbin, S.; Rousseaux, S. Proteomic strategy for the identification of critical actors in reorganization of the post-meiotic male genome. Mol. Hum. Reprod. 2012, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kishigami, S.; Wakayama, S.; Nguyen, V.T.; Wakayama, T. Similar time restriction for intracytoplasmic sperm injection and round spermatid injection into activated oocytes for efficient offspring production. Biol. Reprod. 2004, 70, 1863–1869. [Google Scholar] [CrossRef] [PubMed]
- Pittoggi, C.; Magnano, A.R.; Sciamanna, I.; Giordano, R.; Lorenzini, R.; Spadafora, C. Specific localization of transcription factors in the chromatin of mouse mature spermatozoa. Mol. Reprod. Dev. 2001, 60, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Philpott, A.; Leno, G.H.; Laskey, R.A. Sperm decondensation in xenopus egg cytoplasm is mediated by nucleoplasmin. Cell 1991, 65, 569–578. [Google Scholar] [CrossRef]
- Philpott, A.; Leno, G.H. Nucleoplasmin remodels sperm chromatin in xenopus egg extracts. Cell 1992, 69, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, P.J.; Gambus, A.; Blow, J.J. Preparation and use of xenopus egg extracts to study DNA replication and chromatin associated proteins. Methods 2012, 57, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, J.M.; Vassetzky, Y.; Mechali, M. Analysis of chromatin assembly, chromatin domains, and DNA replication using xenopus systems. In Mapping Protein/DNA Interactions by Cross-Linking; INSERM: Paris, France, 2001. [Google Scholar]
- Risley, M.S.; Eckhardt, R.A. Dissociation and separation of xenopus laevis spermatogenic cells. J. Exp. Zool. 1979, 207, 93–106. [Google Scholar] [CrossRef]
- Teperek, M. Programming of the Paternal Nucleus for Embryonic Development; University of Cambridge: Cambridge, UK, 2014. [Google Scholar]
- Abe, S.; Hiyoshi, H. Synthesis of sperm-specific basic nuclear proteins (sps) in cultured spermatids from xenopus laevis. Exp. Cell Res. 1991, 194, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Hiyoshi, H.; Uno, S.; Yokota, T.; Katagiri, C.; Nishida, H.; Takai, M.; Agata, K.; Eguchi, G.; Abe, S. Isolation of cdna for a xenopus sperm-specific basic nuclear protein (sp4) and evidence for expression of sp4 mrna in primary spermatocytes. Exp. Cell Res. 1991, 194, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Hirosawa, M.; Hoshida, M.; Ishikawa, M.; Toya, T. Mascot: Multiple alignment system for protein sequences based on three-way dynamic programming. Comput. Appl. Biosci. CABIOS 1993, 9, 161–167. [Google Scholar]
- Shechter, D.; Nicklay, J.J.; Chitta, R.K.; Shabanowitz, J.; Hunt, D.F.; Allis, C.D. Analysis of histones in Xenopus laevis. I. A distinct index of enriched variants and modifications exists in each cell type and is remodeled during developmental transitions. J. Biol. Chem. 2009, 284, 1064–1074. [Google Scholar]
- Yang, M.; Li, S.; Liang, G.; Sun, L.; Zhang, X.; Liu, N.; Pang, H.; Miao, S.; Wang, L.; Rao, Z. Crystallization and preliminary crystallographic analysis of rsb-66, a novel round spermatid-specific protein. Acta Crystallogr. D Biol. Crystallogr. 2003, 59, 1853–1855. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hu, T.; Liang, G.; Yang, M.; Zong, S.; Miao, S.; Koide, S.S.; Wang, L. A novel testis protein, RSB-66, interacting with INCA1 (inhibitor of Cdk interacting with cyclin A1). Biochem. Cell Biol. 2008, 86, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Wysocka, J.; Swigut, T.; Milne, T.A.; Dou, Y.; Zhang, X.; Burlingame, A.L.; Roeder, R.G.; Brivanlou, A.H.; Allis, C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 2005, 121, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Lee, H.; Brunzelle, J.S.; Couture, J.F. The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases. Nucleic Acids Res. 2012, 40, 4237–4246. [Google Scholar] [CrossRef] [PubMed]
- Wysocka, J.; Swigut, T.; Xiao, H.; Milne, T.A.; Kwon, S.Y.; Landry, J.; Kauer, M.; Tackett, A.J.; Chait, B.T.; Badenhorst, P.; et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006, 442, 86–90. [Google Scholar] [PubMed]
- Ang, Y.S.; Tsai, S.Y.; Lee, D.F.; Monk, J.; Su, J.; Ratnakumar, K.; Ding, J.; Ge, Y.; Darr, H.; Chang, B.; et al. WDR5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 2011, 145, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Winteringham, L.N.; Kobelke, S.; Williams, J.H.; Ingley, E.; Klinken, S.P. Myeloid leukemia factor 1 inhibits erythropoietin-induced differentiation, cell cycle exit and p27kip1 accumulation. Oncogene 2004, 23, 5105–5109. [Google Scholar] [CrossRef] [PubMed]
- Yoneda-Kato, N.; Kato, J.Y. Shuttling imbalance of MLF1 results in p53 instability and increases susceptibility to oncogenic transformation. Mol. Cell. Biol. 2008, 28, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Yoneda-Kato, N.; Tomoda, K.; Umehara, M.; Arata, Y.; Kato, J.Y. Myeloid leukemia factor 1 regulates p53 by suppressing cop1 via cop9 signalosome subunit 3. EMBO J. 2005, 24, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, I.; Thompson, W.E.; Welch, C.; Thomas, K.; Matthews, R. Prohibitin (PHB) inhibits apoptosis in rat granulosa cells (GCs) through the extracellular signal-regulated kinase 1/2 (ERK1/2) and the Bcl family of proteins. Apoptosis 2013, 18, 1513–1525. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Erdel, F.; Rippe, K. Chromatin remodelling in mammalian cells by iswi-type complexes—Where, when and why? FEBS J. 2011, 278, 3608–3618. [Google Scholar] [CrossRef] [PubMed]
- Poot, R.A.; Bozhenok, L.; van den Berg, D.L.; Hawkes, N.; Varga-Weisz, P.D. Chromatin remodeling by wstf-iswi at the replication site: Opening a window of opportunity for epigenetic inheritance? Cell Cycle 2005, 4, 543–546. [Google Scholar] [CrossRef]
- Tong, J.K.; Hassig, C.A.; Schnitzler, G.R.; Kingston, R.E.; Schreiber, S.L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 1998, 395, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Kikyo, N.; Wade, P.A.; Guschin, D.; Ge, H.; Wolffe, A.P. Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal atpase iswi. Science 2000, 289, 2360–2362. [Google Scholar] [CrossRef] [PubMed]
- Loiodice, I.; Alves, A.; Rabut, G.; van Overbeek, M.; Ellenberg, J.; Sibarita, J.B.; Doye, V. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell 2004, 15, 3333–3344. [Google Scholar] [CrossRef] [PubMed]
- Fontoura, B.M.; Blobel, G.; Matunis, M.J. A conserved biogenesis pathway for nucleoporins: Proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 1999, 144, 1097–1112. [Google Scholar] [CrossRef] [PubMed]
- Fontoura, B.M.; Dales, S.; Blobel, G.; Zhong, H. The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc. Natl. Acad. Sci. USA 2001, 98, 3208–3213. [Google Scholar] [CrossRef] [PubMed]
- Enninga, J.; Levay, A.; Fontoura, B.M. Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol. Cell. Biol. 2003, 23, 7271–7284. [Google Scholar] [CrossRef] [PubMed]
- Rathbone, A.J.; Liddell, S.; Campbell, K.H. Proteomic analysis of early reprogramming events in murine somatic cells incubated with Xenopus laevis oocyte extracts demonstrates network associations with induced pluripotency markers. Cell. Reprogram. 2013, 15, 269–280. [Google Scholar] [PubMed]
- Miyamoto, K.; Nagai, K.; Kitamura, N.; Nishikawa, T.; Ikegami, H.; Binh, N.T.; Tsukamoto, S.; Matsumoto, M.; Tsukiyama, T.; Minami, N.; et al. Identification and characterization of an oocyte factor required for development of porcine nuclear transfer embryos. Proc. Natl. Acad. Sci. USA 2011, 108, 7040–7045. [Google Scholar] [Green Version]
- Novak, S.; Paradis, F.; Savard, C.; Tremblay, K.; Sirard, M.A. Identification of porcine oocyte proteins that are associated with somatic cell nuclei after co-incubation. Biol. Reprod. 2004, 71, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- Rowles, A.; Blow, J.J. Chromatin proteins involved in the initiation of DNA replication. Curr. Opin. Genet. Dev. 1997, 7, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, B.; Faller, D.V. Prohibitin requires Brg-1 and brm for the repression of E2F and cell growth. EMBO J. 2002, 21, 3019–3028. [Google Scholar] [CrossRef] [PubMed]
- VanDemark, A.P.; Blanksma, M.; Ferris, E.; Heroux, A.; Hill, C.P.; Formosa, T. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol. Cell 2006, 22, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Li, Q.; McCullough, L.; Kettelkamp, C.; Formosa, T.; Zhang, Z. Ubiquitylation of FACT by the cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev. 2010, 24, 1485–1490. [Google Scholar] [CrossRef] [PubMed]
- Sible, J.C.; Erikson, E.; Hendrickson, M.; Maller, J.L.; Gautier, J. Developmental regulation of mcm replication factors in xenopus laevis. Curr. Biol. 1998, 8, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Henricksen, L.A.; Carter, T.; Dutta, A.; Wold, M.S. Phosphorylation of human replication protein a by the DNA-dependent protein kinase is involved in the modulation of DNA replication. Nucleic Acids Res. 1996, 24, 3107–3112. [Google Scholar] [CrossRef] [PubMed]
- Bochkarev, A.; Pfuetzner, R.A.; Edwards, A.M.; Frappier, L. Structure of the single-stranded-DNA-binding domain of replication protein a bound to DNA. Nature 1997, 385, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tam, W.L.; Tong, G.Q.; Wu, Q.; Chan, H.Y.; Soh, B.S.; Lou, Y.; Yang, J.; Ma, Y.; Chai, L.; et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 2006, 8, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.H.; Au, K.F.; Leong, D.E.; Foygel, K.; Wong, W.H.; Yao, M.W. An Oct4-Sall4-Nanog network controls developmental progression in the pre-implantation mouse embryo. Mol. Syst. Biol. 2013, 9. [Google Scholar] [CrossRef]
- Elling, U.; Klasen, C.; Eisenberger, T.; Anlag, K.; Treier, M. Murine inner cell mass-derived lineages depend on sall4 function. Proc. Natl. Acad. Sci. USA 2006, 103, 16319–16324. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Hatada, S.; Asashima, M.; Noda, M. HMG-X, a xenopus gene encoding an HMG1 homolog, is abundantly expressed in the developing nervous system. FEBS Lett. 1994, 352, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Ray-Gallet, D.; Quivy, J.P.; Scamps, C.; Martini, E.M.; Lipinski, M.; Almouzni, G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 2002, 9, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Lachner, M.; O’Carroll, D.; Rea, S.; Mechtler, K.; Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001, 410, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.H.; Workman, J.L. The changing faces of HP1: From heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription. Bioessays 2011, 33, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, A.; Hon, G.C.; Jin, F.; Henry, R.E.; Espinosa, J.M.; Ren, B. CBX3 regulates efficient rna processing genome-wide. Genome Res. 2012, 22, 1426–1436. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, R.; Gonzales-Cope, M.; Chronis, C.; Bonora, G.; McKee, R.; Huang, C.; Patel, S.; Lopez, D.; Mishra, N.; Pellegrini, M.; et al. Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1γ in reprogramming to pluripotency. Nat. Cell Biol. 2013, 15, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Wade, P.A.; Gegonne, A.; Jones, P.L.; Ballestar, E.; Aubry, F.; Wolffe, A.P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 1999, 23, 62–66. [Google Scholar] [PubMed]
- Iwano, H.; Nakamura, M.; Tajima, S. Xenopus MBD3 plays a crucial role in an early stage of development. Dev. Biol. 2004, 268, 416–428. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, R.L.; Tosti, L.; Radzisheuskaya, A.; Caballero, I.M.; Kaji, K.; Hendrich, B.; Silva, J.C. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell 2014, 15, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Rais, Y.; Zviran, A.; Geula, S.; Gafni, O.; Chomsky, E.; Viukov, S.; Mansour, A.A.; Caspi, I.; Krupalnik, V.; Zerbib, M.; et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 2013, 502, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Laherty, C.D.; Yang, W.M.; Sun, J.M.; Davie, J.R.; Seto, E.; Eisenman, R.N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 1997, 89, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Hassig, C.A.; Tong, J.K.; Fleischer, T.C.; Owa, T.; Grable, P.G.; Ayer, D.E.; Schreiber, S.L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl. Acad. Sci. USA 1998, 95, 3519–3524. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.C.; Valkov, N.; Yang, W.M.; Gump, J.; Sullivan, D.; Seto, E. Histone deacetylase interacts directly with DNA topoisomerase II. Nat. Genet. 2000, 26, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, S.; Jacques, V.; Rusche, J.R.; Olson, E.N.; Cairns, B.R.; Chandrasekharan, M.B. Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression. Epigenetics Chromatin 2013, 6. [Google Scholar] [CrossRef] [PubMed]
- Newport, J.; Kirschner, M. A major developmental transition in early xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell 1982, 30, 675–686. [Google Scholar]
- Hah, N.; Kolkman, A.; Ruhl, D.D.; Pijnappel, W.W.; Heck, A.J.; Timmers, H.T.; Kraus, W.L. A role for BAF57 in cell cycle-dependent transcriptional regulation by the SWI/SNF chromatin remodeling complex. Cancer Res. 2010, 70, 4402–4411. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.J.; Fairclough, L.; Latinkic, B.V.; Sparrow, D.B.; Mohun, T.J. Xenopus laevis transgenesis by sperm nuclear injection. Nat. Protoc. 2006, 1, 2195–2203. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Teperek, M.; Miyamoto, K.; Simeone, A.; Feret, R.; Deery, M.J.; Gurdon, J.B.; Jullien, J. Sperm and Spermatids Contain Different Proteins and Bind Distinct Egg Factors. Int. J. Mol. Sci. 2014, 15, 16719-16740. https://doi.org/10.3390/ijms150916719
Teperek M, Miyamoto K, Simeone A, Feret R, Deery MJ, Gurdon JB, Jullien J. Sperm and Spermatids Contain Different Proteins and Bind Distinct Egg Factors. International Journal of Molecular Sciences. 2014; 15(9):16719-16740. https://doi.org/10.3390/ijms150916719
Chicago/Turabian StyleTeperek, Marta, Kei Miyamoto, Angela Simeone, Renata Feret, Michael J. Deery, John B. Gurdon, and Jerome Jullien. 2014. "Sperm and Spermatids Contain Different Proteins and Bind Distinct Egg Factors" International Journal of Molecular Sciences 15, no. 9: 16719-16740. https://doi.org/10.3390/ijms150916719




