MicroRNA Expression Profiling of Lactating Mammary Gland in Divergent Phenotype Swine Breeds
Abstract
:1. Introduction
2. Results
2.1. Determination of Porcine Mammary Gland
2.2. Analysis of Sequencing Data



| Category | Jinhua Mammary Gland | Yorkshire Mammary Gland |
|---|---|---|
| Total miRNAs | 3364 | 2830 |
| Known miRNAs | 406 | 406 |
| Conserved miRNAs | 135 | 138 |
| Predicted miRNAs | 2823 | 2286 |
| Mapped to known miRs of selected species and genome | 552 | 587 |
| Mapped to known miRs and miRs of selected species but unmapped to genome | 571 | 595 |
| Unmapped to known miRs but mapped to genome and within hairpins | 1700 | 1104 |
2.3. Comparison of Expression Levels of miRNAs in RX_J and RX_Y

2.4. The Distribution of miRNA on the Porcine Genome

2.5. The Conservation Profile of Conserved Porcine miRNAs
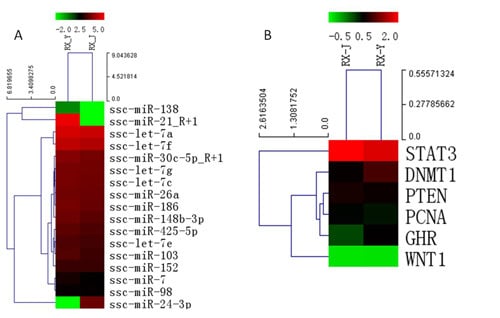
2.6. Identification of Differential Expression Patterns of miRNA in Porcine Mammary Gland

| miRNA Name | Sequence (5'–3') | Regulation | p-Value |
|---|---|---|---|
| ssc-miR-21 | TAGCTTATCAGACTGATGTTGA | Up | 0 |
| ssc-miR-148b-3p | UCAGUGCAUCAGAACUUUGU | Up | 0 |
| ssc-miR-92a | TATTGCACTTGTCCCGGCCTGT | Up | 3.4 × 10−135 |
| ssc-miR-423-3p | AGCUCGGCUGAGGCCCCUCAGU | Up | 1.1 × 10−154 |
| ssc-miR-26 | TTCAAGTAATCCAGGATAGGCT | Down | 4 × 10−155 |
| ssc-miR-24-3p | UGGCUCAGUUCAGCAGGAACAG | Down | 1.37 × 10−81 |
| ssc-miR-181a | AACAUUCAACGCUGUCGGUGAGUU | Down | 1.67 × 10−61 |
| ssc-miR-151-5p | UCGAGGAGCUCAGUCUAGU | Down | 6.9 × 10−4 |
2.7. Gene Ontology (GO) Enrichment Analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of Target Genes

2.8. Regulation of Lactation Genes by miRNAs

2.9. Validation of miRNA Expression with Quantitative RT-PCR

3. Discussion
4. Experimental Section
4.1. Ethics Statement
4.2. Sample Collection and RNA Extraction
4.3. Small RNA Library Construction and Sequencing
4.4. Data Processing
4.5. Analysis of Differential Expressed miRNAs
4.6. The Prediction of Target Genes of miRNAs
4.7. Histologic Examination
4.8. Immunohistochemical Analysis and Immunofluorescence Assay
4.9. Quantitative RT-PCR Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Galio, L.; Droineau, S.; Yeboah, P.; Boudiaf, H.; Bouet, S.; Truchet, S.; Devinoy, E. MicroRNA in the ovine mammary gland during early pregnancy: Spatial and temporal expression of miR-21, miR-205, and miR-200. Physiol. Genomics 2013, 45, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Silveri, L.; Tilly, G.; Vilotte, J.; le Provost, F. MicroRNA involvement in mammary gland development and breast cancer. Reprod. Nutr. Dev. 2006, 46, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, H.; Jin, X.; Lo, L.; Liu, J. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genomics 2012, 13, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Manjarin, R.; Steibel, J.P.; Kirkwood, R.N.; Taylor, N.P.; Trottier, N.L. Transcript abundance of hormone receptors, mammalian target of rapamycin pathway-related kinases, insulin-like growth factor I, and milk proteins in porcine mammary tissue. J. Anim. Sci. 2012, 90, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Yoon, K.; Cho, K.; Kim, K.; Jung, H. Expression of miR-206 during the initiation of mammary gland development. Cell Tissue Res. 2013, 353, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Ucar, A.; Vafaizadeh, V.; Jarry, H.; Fiedler, J.; Klemmt, P.A.B.; Thum, T.; Groner, B.; Chowdhury, K. miR-212 and miR-132 are required for epithelial stromal interactions necessary for mouse mammary gland development. Nat. Genet. 2010, 42, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Avril-Sassen, S.; Goldstein, L.D.; Stingl, J.; Blenkiron, C.; le Quesne, J.; Spiteri, I.; Karagavriilidou, K.; Watson, C.J.; Tavare, S.; Miska, E.A.; et al. Characterisation of microRNA expression in post-natal mouse mammary gland development. BMC Genomics 2009, 10, 548. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, C.; Li, Q.; Gao, X. MiR-15a decreases bovine mammary epithelial cell viability and lactation and regulates growth hormone receptor expression. Molecules 2012, 17, 12037–12048. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Moisá, S.; Khan, M.J.; Wang, J.; Bu, D.; Loor, J.J. MicroRNA expression patterns in the bovine mammary gland are affected by stage of lactation. J. Dairy Sci. 2012, 95, 6529–6535. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lan, X.; Guo, W.; Sun, J.; Huang, Y.; Wang, J.; Huang, T.; Lei, C.; Fang, X.; Chen, H. Comparative transcriptome profiling of dairy goat microRNAs from dry period and peak lactation mammary gland tissues. PLoS One 2012, 7, 52349–52388. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.F.; Liston, A. MicroRNA in the immune system, microRNA as an immune system. Immunology 2009, 127, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Margaret, C.N.; Thomas, B.M.; Isabel, F. Hormonal regulation of mammary differentiation and milk secretion. J. Mammary Gland Biol. Neoplasia 2002, 7, 49–66. [Google Scholar] [CrossRef] [PubMed]
- Chomwisarutkun, K.; Murani, E.; Ponsuksili, S.; Wimmers, K. Gene expression analysis of mammary tissue during fetal bud formation and growth in two pig breeds—Indications of prenatal initiation of postnatal phenotypic differences. BMC Dev. Biol. 2012, 12. [Google Scholar] [CrossRef]
- Hurley, W.L. Mammary gland growth in the lactating sow. Livest. Prod. Sci. 2001, 70, 149–157. [Google Scholar] [CrossRef]
- Vanklompenberg, M.K.; Manjarin, R.; Trott, J.F.; McMicking, H.F.; Hovey, R.C. Late gestational hyperprolactinemia accelerates mammary epithelial cell differentiation that leads to increased milk yield. J. Anim. Sci. 2013, 91, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol. Genomics 2007, 29, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, Q. Identification of differentially expressed microRNAs during the development of Chinese murine mammary gland. J. Genet. Genomics 2007, 34, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Li, M.; Wang, T.; Liang, Y.; Zhong, Z.; Wang, X.; Zhou, Q.; Chen, L.; Lang, Q.; He, Z.; et al. Lactation-related microRNA expression profiles of porcine breast milk exosomes. PLoS One 2012, 7, e43691. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Mao, S.; Shah, S.; Gorski, D.H.; Krawetz, S.A.; Sloane, B.F.; Mattingly, R.R. Next-generation sequencing: A powerful tool for the discovery of molecular markers in breast ductal carcinoma in situ. Expert Rev. Mol. Diagn. 2013, 13, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Zhang, Z.; Yuan, Z.; Lo, L.J.; Chen, J.; Wang, Y.; Peng, J. Distinctive genes determine different intramuscular fat and muscle fiber ratios of the longissimus dorsi muscles in Jinhua and landrace pigs. PLoS One 2013, 8, e53181. [Google Scholar] [CrossRef] [PubMed]
- Uimari, P.; Sironen, A.; Sevon-Aimonen, M.L. Evidence for three highly significant QTL for meat quality traits in the Finnish Yorkshire pig breed. J. Anim. Sci. 2013, 91, 2001–2011. [Google Scholar] [CrossRef] [PubMed]
- Burgos, S.A.; Dai, M.; Cant, J.P. Nutrient availability and lactogenic hormones regulate mammary protein synthesis through the mammalian target of rapamycin signaling pathway. J. Dairy Sci. 2010, 93, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Sobolewska, A.; Gajewska, M.; Zarzynska, J.; Gajkowska, B.; Motyl, T. IGF-I, EGF, and sex steroids regulate autophagy in bovine mammary epithelial cells via the mTOR pathway. Eur. J. Cell Biol. 2009, 88, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Haneda, S.; Imakawa, K.; Sakai, S.; Nagaoka, K. A microRNA, miR-101a, controls mammary gland development by regulating cyclooxygenase-2 expression. Differentiation 2009, 77, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Li, Q.; Feng, L.; Ding, W. MiR-126–3p regulates progesterone receptors and involves development and lactation of mouse mammary gland. Mol. Cell. Biochem. 2011, 355, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, R.C.; Bierie, B.; Zhao, L.; Raz, R.; Levy, D.; Hennighausen, L. Deletion of Stat3 blocks mammary gland involution and extends functional competence of the secretory epithelium in the absence of lactogenic stimuli. Endocrinology 2002, 143, 3641–3650. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Song, Y.X.; Wang, Z.N. The microRNA-148/152 family: Multi-faceted players. Mol. Cancer 2013, 12, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Robinson, G.W.; Lesche, R.; Martinez-Diaz, H.; Jiang, Z.; Rozengurt, N.; Wagner, K.U.; Wu, D.C.; Lane, T.F.; Liu, X.; et al. Conditional loss of PTEN leads to precocious development and neoplasia in the mammary gland. Development 2002, 129, 4159–4170. [Google Scholar] [PubMed]
- Paunesku, T.; Mittal, S.; Protic, M.; Oryhon, J.; Korolev, S.V.; Joachimiak, A.; Woloschak, G.E. Proliferating cell nuclear antigen (PCNA): Ringmaster of the genome. Int. J. Radiat. Biol. 2001, 77, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Brisken, C.; O’Malley, B. Hormone action in the mammary gland. Cold Spring Harb. Perspect. Biol. 2010, 2, a3178. [Google Scholar] [CrossRef]
- Chu, E.Y.; Hens, J.; Andl, T.; Kairo, A.; Yamaguchi, T.P.; Brisken, C.; Glick, A.; Wysolmerski, J.J.; Millar, S.E. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development 2004, 131, 4819–4829. [Google Scholar] [CrossRef] [PubMed]
- RNAfold WebServer. Available online: http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi (accessed on 26 May 2013).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, J.; Zhao, J.-S.; Shen, Y.-F.; Mao, H.-G.; Xu, N.-Y. MicroRNA Expression Profiling of Lactating Mammary Gland in Divergent Phenotype Swine Breeds. Int. J. Mol. Sci. 2015, 16, 1448-1465. https://doi.org/10.3390/ijms16011448
Peng J, Zhao J-S, Shen Y-F, Mao H-G, Xu N-Y. MicroRNA Expression Profiling of Lactating Mammary Gland in Divergent Phenotype Swine Breeds. International Journal of Molecular Sciences. 2015; 16(1):1448-1465. https://doi.org/10.3390/ijms16011448
Chicago/Turabian StylePeng, Jing, Jun-Sheng Zhao, Yi-Fei Shen, Hai-Guang Mao, and Ning-Ying Xu. 2015. "MicroRNA Expression Profiling of Lactating Mammary Gland in Divergent Phenotype Swine Breeds" International Journal of Molecular Sciences 16, no. 1: 1448-1465. https://doi.org/10.3390/ijms16011448