Development and Application of a Label-Free Fluorescence Method for Determining the Composition of Gold Nanoparticle–Protein Conjugates
Abstract
:1. Introduction
2. Results and Discussion
2.1. Measurement of the Dimensions of the Gold Nanoparticles

2.2. Using the Protein’s Intrinsic Fluorescence to Determine the Composition of the Protein-Gold Nanoparticle Conjugates
Cconjugated = C0 − Cunbound = C0 (F0 − F)/F0
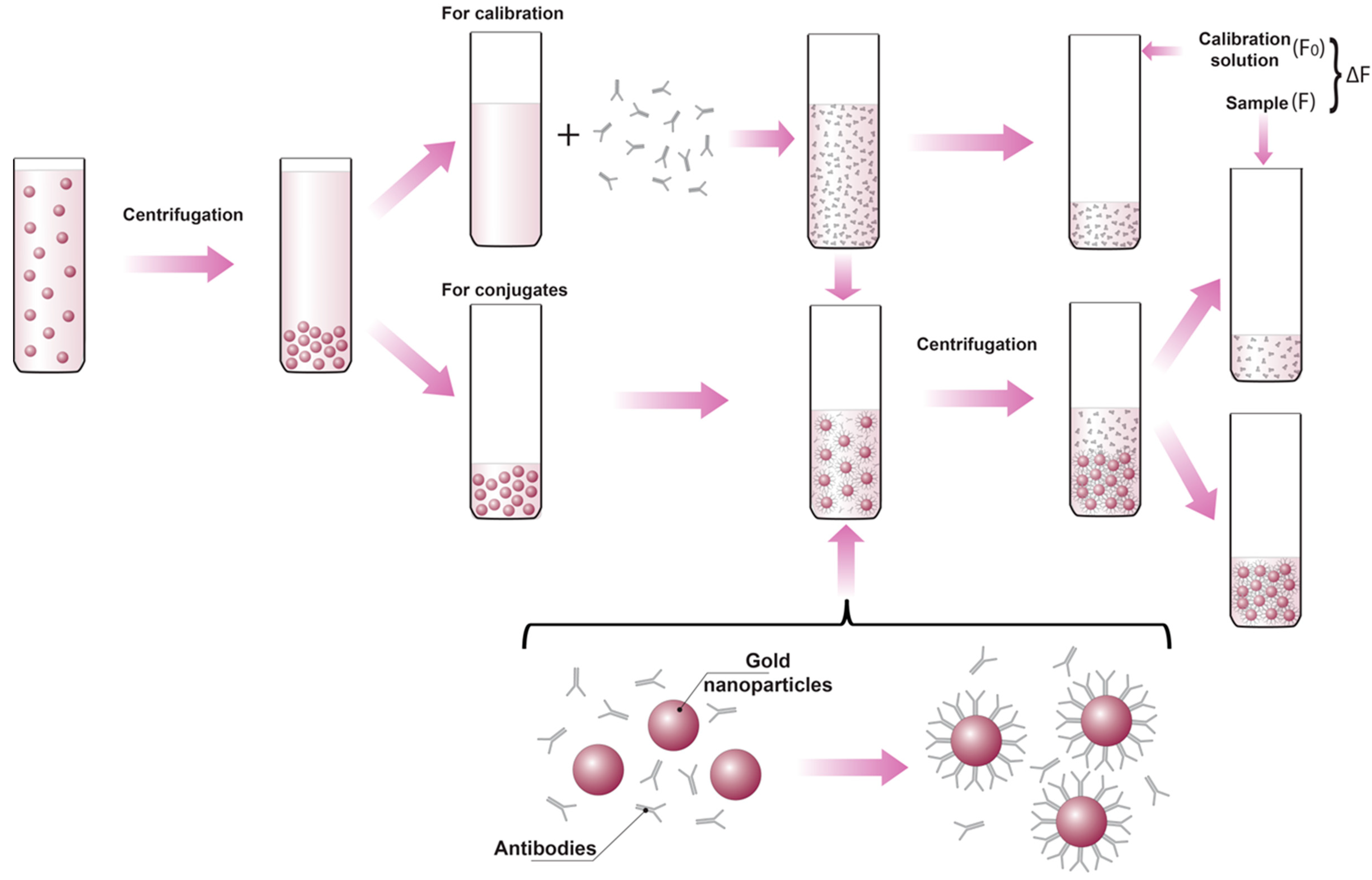
2.3. Determination of the Concentration of the Gold Nanoparticles
2.4. Determination of the Amount of Protein Adsorbed on the Gold Nanoparticles


2.5. Determination of the Equilibrium Dissociation Constants of the Protein-Nanoparticle Interaction and the Number of Sorption Sites

| Protein | MW, kD | 1/MW, kD−1 | Kd(1), nM | Kd(2), nM | N |
|---|---|---|---|---|---|
| IgG | 150 | 0.00667 | 4 | 30 | 52 |
| BSA | 66 | 0.01515 | 6 | 76 | 90 |
| Protein G | 26 | 0.03846 | 10 | 175 | 500 |
| STI | 20 | 0.05 | 15 | 100 | 550 |


3. Materials and Methods
3.1. Reagents
3.2. Preparation of Gold Nanoparticles
3.3. Transmission Electron Microscopy
3.4. Dynamic Light Scattering
3.5. Preparation of the Gold Nanoparticle-Protein Conjugates
3.6. Fluorescence Measurements
3.7. Three-Dimensional Modeling the Gold Nanoparticle-Protein Conjugates
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sperling, R.A.; Rivera, P.; Zhang, F.; Zanella, M.; Parak, W.J. Biological applications of gold nanoparticles. Chem. Soc. Rev. 2008, 37, 1896–1908. [Google Scholar] [CrossRef] [PubMed]
- Boisselier, E.; Astruc, D. Gold nanoparticles in nanomedicine: Preparations, imaging, diagnostics, therapies and toxicity. Chem. Soc. Rev. 2009, 38, 1759–1782. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.W.; Yong, K.T.; Roy, I.; Dinh, X.Q.; Yu, X.; Luan, F. A review on functionalized gold nanoparticles for biosensing applications. Plasmonics 2011, 6, 491–506. [Google Scholar] [CrossRef]
- Saha, K.; Agasti, S.S.; Kim, C.; Li, X.N.; Rotello, V.M. Gold nanoparticles in chemical and biological sensing. Chem. Rev. 2012, 112, 2739–2779. [Google Scholar] [CrossRef] [PubMed]
- Dreaden, E.C.; Alkilany, A.M.; Huang, X; Murphy, C.J.; el-Sayed, M.A. The golden age: Gold nanoparticles for biomedicine. Chem. Soc. Rev. 2012, 41, 2740–2779. [Google Scholar] [CrossRef]
- Dykman, L.; Khlebtsov, N. Gold nanoparticles in biology and medicine: Recent advances and prospects. Acta Naturae 2011, 3, 34–55. [Google Scholar] [PubMed]
- Sapsford, K.E.; Algar, W.R.; Berti, L; Gemmill, K.B.; Casey, B.J.; Oh, E.; Stewart, M.H.; Medintz, I.L. Functionalizing nanoparticles with biological molecules: Developing chemistries that facilitate nanotechnology. Chem. Rev. 2013, 113, 1904–2074. [Google Scholar] [CrossRef]
- Salata, O.V. Applications of nanoparticles in biology and medicine. J. Nanobiotechnol. 2004, 2. [Google Scholar] [CrossRef] [Green Version]
- Casals, E.; Pfaller, T.; Duschl, A.; Oostingh, G.J.; Puntes, V. Time evolution of the nanoparticle protein corona. ACS Nano 2010, 4, 3623–3632. [Google Scholar] [CrossRef] [PubMed]
- Tsai, D.H.; DelRio, F.W.; Keene, A.M.; Tyner, K.M.; MacCuspie, R.I.; Cho, T.J.; Zachariah, M.R.; Hackley, V.A. Adsorption and conformation of serum albumin protein on gold nanoparticles investigated using dimensional measurements and in situ spectroscopic methods. Langmuir 2011, 27, 2464–2477. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Medina, S.; McDonough, S.; Swanglap, P; Landes, C.F.; Link, S. In situ measurement of bovine serum albumin interaction with gold nanospheres. Langmuir 2012, 28, 9131–9139. [Google Scholar]
- Lacerda, S.H.D.P.; Park, J.J.; Meuse, C.; Pristinski, D.; Becker, M.L.; Karim, A.; Douglas, J.F. Interaction of gold nanoparticles with common human blood proteins. ACS Nano 2010, 4, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Joshi, P.; Shanker, V; Ansari, Z.A.; Singh, S.P.; Chakrabarti, P. Contrasting effect of gold nanoparticles and nanorods with different surface modifications on the structure and activity of bovine serum albumin. Langmuir 2011, 27, 7722–7731. [Google Scholar] [CrossRef]
- De Roe, C.; Courtoy, P.J.; Baudhuin, P. A model of protein-colloidal gold interaction. J. Histochem. Cytochem. 1987, 35, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Ghitescu, L.; Bendayan, M. Immunolabeling efficiency of protein A-gold complexes. J. Histochem. Cytochem. 1990, 38, 1523–1530. [Google Scholar] [CrossRef] [PubMed]
- Naveenraj, S.; Anandan, S.; Kathiravan, A.; Renganathan, R.; Ashokkumar, M. The interaction of sonochemically synthesized gold nanoparticles with serum albumins. J. Pharm. Biomed. Anal. 2010, 53, 804–810. [Google Scholar] [CrossRef]
- Brewer, S.H.; Glomm, W.R.; Johnson, M.C.; Knag, M.K.; Franzen, S. Probing BSA binding to citrate-coated gold nanoparticles and surfaces. Langmuir 2005, 21, 9303–9307. [Google Scholar] [CrossRef] [PubMed]
- Sen, T.; Haldar, K.K.; Patra, A. Au nanoparticle-based surface energy transfer probe for conformational changes of BSA protein. J. Phys. Chem. C 2008, 112, 17945–17951. [Google Scholar] [CrossRef]
- Sapsford, K.E.; Tyner, K.M.; Dair, B.J.; Deschamps, J.R.; Medintz, I.L. Analyzing nanomaterial bioconjugates: A review of current and emerging purification and characterization techniques. Anal. Chem. 2011, 83, 4453–4488. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Narsimhan, G. Characterization of secondary and tertiary conformational changes of β-lactoglobulin on silica nanoparticle surfaces. Langmuir 2008, 24, 4989–4998. [Google Scholar] [CrossRef] [PubMed]
- Naveenraj, S.; Anandan, S. Binding of serum albumins with bioactive substances—Nanoparticles to drugs. J. Photochem. Photobiol. C 2013, 14, 53–71. [Google Scholar] [CrossRef]
- Shang, L.; Wang, Y.; Jiang, J.; Dong, S. pH-dependent protein conformational changes in albumin: Gold nanoparticle bioconjugates: A spectroscopic study. Langmuir 2007, 23, 2714–2721. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Wang, J.; Jasinski, J.; Achilefu, J. Fluorescent manipulation by gold nanoparticles: From complete quenching to extensive enhancement. J. Nanobiotechnol. 2011, 9. [Google Scholar] [CrossRef]
- Goward, C.R.; Murphy, J.P.; Atkinson, T.; Barstow, D.A. Expression and purification of a truncated recombinant streptococcal protein G. Biochem. J. 1990, 267, 171–177. [Google Scholar] [PubMed]
- O’Connell, T.X.; Horita, T.J.; Kasravi, B. Understanding and interpreting serum protein electrophoresis. Am. Fam. Phys. 2005, 71, 105–112. [Google Scholar]
- Dobrovolskaia, M.A.; Patri, A.K.; Zheng, J.; Clogston, J.D.; Ayub, N.; Aggarwal, P.; Neun, B.W.; Hall, J.B.; McNeil, S.E. Interaction of colloidal gold nanoparticles with human blood: Effects on particle size and analysis of plasma protein binding profiles. Nanomedicine 2009, 5, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer Science and Business Media: New York, NY, USA, 2010; p. 954. [Google Scholar]
- Engelborghs, Y. Correlating protein structure and protein fluorescence. J. Fluoresc. 2003, 13, 9–16. [Google Scholar] [CrossRef]
- Vivian, J.T.; Callis, P.R. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 2001, 80, 2093–2109. [Google Scholar] [CrossRef] [PubMed]
- Scatchard, G. The Attractions of proteins for small molecules and ions. Ann. N. Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- Dykman, L.; Khlebtsov, N. Gold nanoparticles in biomedical applications: Recent advances and perspectives. Chem. Soc. Rev. 2012, 41, 2256–2282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Moustafa, Y.; Huo, Q. Different interaction modes of biomolecules with citrate-capped gold nanoparticles. ACS Appl. Mater. Interfaces 2014, 6, 21184–21192. [Google Scholar] [CrossRef] [PubMed]
- Saptarshi, S.R.; Duschl, A.; Lopata, A.L. Interaction of nanoparticles with proteins: Relation to bio-reactivity of the nanoparticle. J. Nanobiotechnol. 2013, 11. [Google Scholar] [CrossRef]
- Wang, A.; Vangala, K.; Vo, T.; Zhang, D.; Fitzkee, N.C. A three-step model for protein-gold nanoparticle adsorption. J. Phys. Chem. C 2014, 118, 8134–8142. [Google Scholar] [CrossRef]
- Chaudhary, A.; Gupta, A.; Khan, S.; Nandi, C.K. Morphological effect of gold nanoparticles on the adsorption of bovine serum albumin. Phys. Chem. Chem. Phys. 2014, 16, 20471–20482. [Google Scholar] [CrossRef] [PubMed]
- Khlebtsov, N.G.; Bogatyrev, V.A.; Khlebtsov, B.N.; Dykman, L.A.; Englebienne, P. A multilayer model for gold nanoparticle bioconjugates: Application to study of gelatin and human IgG adsorption using extinction and light scattering spectra and the dynamic light scattering method. Colloid J. 2003, 65, 622–635. [Google Scholar] [CrossRef]
- Nienhaus, G.U.; Maffre, P.; Nienhaus, K. Studying the protein corona on nanoparticles by FCS. Methods Enzymol. 2013, 519, 115–137. [Google Scholar] [PubMed]
- Frens, G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nat. Phys. Sci. 1973, 241, 20–22. [Google Scholar] [CrossRef]
- Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. Factors influencing the detection limit of the lateral-flow sandwich immunoassay: A case study with potato virus X. Anal. Bioanal. Chem. 2012, 403, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sotnikov, D.V.; Zherdev, A.V.; Dzantiev, B.B. Development and Application of a Label-Free Fluorescence Method for Determining the Composition of Gold Nanoparticle–Protein Conjugates. Int. J. Mol. Sci. 2015, 16, 907-923. https://doi.org/10.3390/ijms16010907
Sotnikov DV, Zherdev AV, Dzantiev BB. Development and Application of a Label-Free Fluorescence Method for Determining the Composition of Gold Nanoparticle–Protein Conjugates. International Journal of Molecular Sciences. 2015; 16(1):907-923. https://doi.org/10.3390/ijms16010907
Chicago/Turabian StyleSotnikov, Dmitriy V., Anatoly V. Zherdev, and Boris B. Dzantiev. 2015. "Development and Application of a Label-Free Fluorescence Method for Determining the Composition of Gold Nanoparticle–Protein Conjugates" International Journal of Molecular Sciences 16, no. 1: 907-923. https://doi.org/10.3390/ijms16010907







