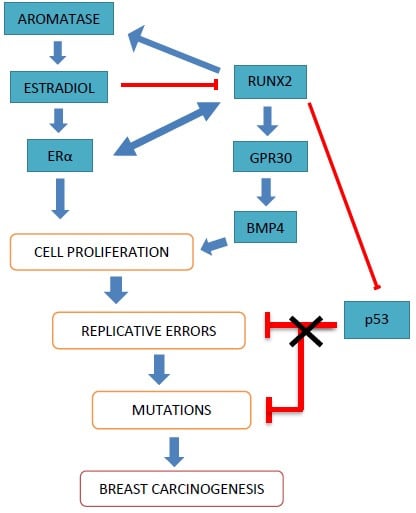
Role of RUNX2 in Breast Carcinogenesis
Abstract
:1. Molecular Aspects of Breast Cancer
1.1. Hormone-Dependent Cancers
1.2. Role of Estrogen in Breast Carcinogenesis
1.3. Estrogen Receptors in Breast Cancer
2. RUNX2—A Transcription Factor and the Bone Growth Master Regulator
2.1. The Structure of RUNX2 Gene and Protein
2.2. RUNX2 in Physiology
2.3. Multipathway Regulation of RUNX2

3. Involvement of RUNX2 in DNA Damage Response and Cancer Transformation
3.1. The Cellular DNA Damage Response
3.2. Involvement of RUNX2 in DDR
3.3. Role of RUNX2 in Cancer

4. Role of RUNX2 in Breast Cancer
4.1. Cooperative Action of RUNX2 and Estrogen Signaling in Bone Homeostasis

4.2. Interactions of RUNX2 and Estrogen in Breast Cancer
4.2.1. Estrogen and Estrogen Receptors

4.2.2. HER1 and HER2 Receptors
4.2.3. Cyclin D1
4.2.4. Matrix Metalloproteinases (MMPs)
5. Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Brown, S.B.; Hankinson, S.E. Endogenous estrogens and the risk of breast, endometrial, and ovarian cancers. Steroids 2015, 99, 8–10. [Google Scholar] [CrossRef] [PubMed]
- Jemal, A.; Tiwari, R.C.; Murray, T.; Ghafoor, A.; Samuels, A.; Ward, E.; Feuer, E.J.; Thun, M.J. Cancer statistics. CA: Cancer J. Clin. 2004, 54, 8–29. [Google Scholar]
- Hapangama, D.K.; Kamal, A.M.; Bulmer, J.N. Estrogen receptor β: The guardian of the endometrium. Hum. Reprod. Update 2015, 21, 174–193. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, V.W.; Pike, M.C.; Karageorgi, S.; Deming, S.L.; Anderson, K.; Bernstein, L.; Brinton, L.A.; Cai, H.; Cerhan, J.R.; Cozen, W.; et al. Age at last birth in relation to risk of endometrial cancer: Pooled analysis in the epidemiology of endometrial cancer consortium. Am. J. Epidemiol. 2012, 176, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.R.; Freiman, R.N. Estrogen signaling crosstalk: Implications for endocrine resistance in ovarian cancer. J. Steroid Biochem. Mol. Biol. 2014, 143, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.G.; Zeng, Q.; Tse, G.M. Estrogen and its receptors in cancer. Med. Res. Rev. 2008, 28, 954–974. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, J.M.; Vermeulen, A. The decline of androgen levels in elderly men and its clinical and therapeutic implications. Endocr. Rev. 2005, 26, 833–876. [Google Scholar] [CrossRef] [PubMed]
- Hartman, J.; Ström, A.; Gustafsson, J.Å. Current concepts and significance of estrogen receptor β in prostate cancer. Steroids 2012, 77, 1262–1266. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2012. CA: Cancer J. Clin. 2012, 62, 10–29. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA: Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Love, R.R.; Philips, J. Oophorectomy for breast cancer: History revisited. J. Natl. Cancer Inst. 2002, 94, 1433–1434. [Google Scholar] [CrossRef] [PubMed]
- Creasman, W.T. Estrogen and cancer. Gynecol. Oncol. 2002, 86, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ascenzi, P.; Bocedi, A.; Marino, M. Structure-function relationship of estrogen receptor α and β: Impact on human health. Mol. Asp. Med. 2006, 27, 299–402. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, R.J.; Likis, F.E. Estrogen: Physiology, pharmacology, and formulations for replacement therapy. J. Midwifery Womens Health 2002, 47, 130–138. [Google Scholar] [CrossRef]
- Dey, P.; Barros, R.P.; Warner, M.; Ström, A.; Gustafsson, J.Å. Insight into the mechanisms of action of estrogen receptor β in the breast, prostate, colon, and CNS. J. Mol. Endocrinol. 2013, 51, T61–T74. [Google Scholar] [CrossRef] [PubMed]
- Bulun, S.E.; Lin, Z.; Imir, G.; Amin, S.; Demura, M.; Yilmaz, B.; Martin, R.; Utsunomiya, H.; Thung, S.; Gurates, B.; et al. Regulation of aromatase expression in estrogen-responsive breast and uterine disease: From bench to treatment. Pharmacol. Rev. 2005, 57, 359–383. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Shen, Y.; Li, R. Estrogen synthesis and signaling pathways during aging: From periphery to brain. Trends Mol. Med. 2013, 19, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, S.; Gustafsson, J.Å. Estrogen receptors: Therapies targeted to receptor subtypes. Clin. Pharmacol. Ther. 2011, 89, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, S.; Mäkelä, S.; Treuter, E.; Tujague, M.; Thomsen, J.; Andersson, G.; Enmark, E.; Pettersson, K.; Warner, M.; Gustafsson, J.A. Mechanisms of estrogen action. Physiol. Rev. 2001, 81, 1535–1565. [Google Scholar] [PubMed]
- Cenni, B.; Picard, D. Ligand-independent activation of steroid receptors: New roles for old players. Trends Endocrinol. Metab. 1999, 10, 41–46. [Google Scholar] [CrossRef]
- Pearce, S.T.; Jordan, V.C. The biological role of estrogen receptors α and β in cancer. Crit. Rev. Oncol. Hematol. 2004, 50, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Levin, E.R. Integration of the extranuclear and nuclear actions of estrogen. Mol. Endocrinol. 2005, 19, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Le Romancer, M.; Poulard, C.; Cohen, P.; Sentis, S.; Renoir, J.M.; Corbo, L. Cracking the estrogen receptor’s posttranslational code in breast tumors. Endocr. Rev. 2011, 32, 597–622. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Inoue, S. Estrogen receptors and their downstream targets in cancer. Arch. Histol. Cytol. 2004, 67, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Shanle, E.K.; Xu, W. Selectively targeting estrogen receptors for cancer treatment. Adv. Drug. Deliv. Rev. 2010, 62, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Cai, Q.; Felty, Q.; Narayan, S. Estrogen-induced generation of reactive oxygen and nitrogen species, gene damage, and estrogen-dependent cancers. J. Toxicol. Environ. Health B Crit. Rev. 2007, 10, 235–257. [Google Scholar] [CrossRef] [PubMed]
- Zumoff, B. Does postmenopausal estrogen administration increase the risk of breast cancer? Contributions of animal, biochemical, and clinical investigative studies to a resolution of the controversy. Proc. Soc. Exp. Biol. Med. 1998, 217, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, A.B.; Lerner, M.R.; Lightfoot, S.A.; Wilkerson, K.B.; Hanas, J.S.; McCay, P.B.; Brackett, D.J. Prevention of DMBA-induced rat mammary carcinomas comparing leuprolide, oophorectomy, and tamoxifen. Breast Cancer Res. Treat. 1998, 47, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Yue, W.; Yager, J.D.; Wang, J.P.; Jupe, E.R.; Santen, R.J. Estrogen receptor-dependent and independent mechanisms of breast cancer carcinogenesis. Steroids 2013, 78, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Key, T.; Appleby, P.; Barnes, I.; Reeves, G. Endogenous Hormones and Breast Cancer Collaborative Group. Endogenous sex hormones and breast cancer in postmenopausal women: Reanalysis of nine prospective studies. J. Natl. Cancer Inst. 2002, 94, 606–616. [Google Scholar] [PubMed]
- Kaaks, R.; Rinaldi, S.; Key, T.J.; Berrino, F.; Peeters, P.H.; Biessy, C.; Dossus, L.; Lukanova, A.; Bingham, S.; Khaw, K.T.; et al. Postmenopausal serum androgens, oestrogens and breast cancer risk: The European prospective investigation into cancer and nutrition. Endocr. Relat. Cancer 2005, 12, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Woollard, A. RUNX factors in development: Lessons from invertebrate model systems. Blood Cells Mol. Dis. 2009, 43, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.S.; Ito, K.; Ito, Y. RUNX family: Regulation and diversification of roles through interacting proteins. Int. J. Cancer 2013, 132, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Q.; Crute, B.E.; Melnikova, I.N.; Keller, S.R.; Speck, N.A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol. 1993, 13, 3324–3339. [Google Scholar] [PubMed]
- Gergen, J.P.; Butler, B.A. Isolation of the Drosophila segmentation gene runt and analysis of its expression during embryogenesis. Genes Dev. 1988, 2, 1179–1193. [Google Scholar] [CrossRef] [PubMed]
- Kaminker, J.S.; Singh, R.; Lebestky, T.; Yan, H.; Banerjee, U. Redundant function of runt domain binding partners, Big brother and Brother, during Drosophila development. Development 2001, 128, 2639–2648. [Google Scholar] [PubMed]
- Nimmo, R.; Woollard, A. Worming out the biology of RUNX. Dev. Biol. 2008, 313, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.M., Jr. Perspectives on RUNX genes: An update. Am. J. Med. Genet. A 2009, 149, 2629–2646. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.C.; Lee, Y.H. Phosphorylation, acetylation and ubiquitination: The molecular basis of RUNX regulation. Gene 2006, 366, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Terry, A.; Kilbey, A.; Vaillant, F.; Stewart, M.; Jenkins, A.; Cameron, E.; Neil, J.C. Conservation and expression of an alternative 31 exon of RUNX2 encoding a novel proline-rich C-terminal domain. Gene 2004, 336, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Levanon, D.; Groner, Y. Structure and regulated expression of mammalian RUNX genes. Oncogene 2004, 23, 4211–4219. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Shiga, T.; Ito, Y. RUNX transcription factors in neuronal development. Neural Dev. 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.H.; Choi, J.Y. Interrelationship of RUNX2 and estrogen pathway in skeletal tissues. BMB Rep. 2011, 44, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of skeletal development by the RUNX family of transcription factors. J. Cell. Biochem. 2005, 95, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.; Stricker, S.; Wiecha, U.; Stiege, A.; Panopoulou, G.; Podsiadlowski, L.; Poustka, A.J.; Dieterich, C.; Ehrich, S.; Suvorova, J.; et al. Evolution of a core gene network for skeletogenesis in chordates. PLoS Genet. 2008, 4, e1000025. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010, 339, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.H.; Jin, J.S.; Kim, H.N.; Kang, S.M.; Liu, J.C.; Lengner, C.J.; Otto, F.; Mundlos, S.; Stein, J.L.; van Wijnen, A.J.; et al. Expression of RUNX2 transcription factor in non-skeletal tissues, sperm and brain. J. Cell. Physiol. 2008, 217, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Durst, K.L.; Hiebert, S.W. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene 2004, 23, 4220–4224. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, T.M.; Jensen, E.D.; Westendorf, J.J. RUNX2: A master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res. C Embryo Today 2005, 75, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Westendorf, J.J. Transcriptional co-repressors of RUNX2. J. Cell. Biochem. 2006, 98, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Zambotti, A.; Makhluf, H.; Shen, J.; Ducy, P. Characterization of an osteoblast-specific enhancer element in the CBFA1 gene. J. Biol. Chem. 2002, 277, 41497–41506. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Jiang, D.; Thomas, P.; Benson, M.D.; Guan, K.; Karsenty, G.; Franceschi, R.T. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, CBFA1. J. Biol. Chem. 2000, 275, 4453–4459. [Google Scholar] [CrossRef] [PubMed]
- Fujita, T.; Azuma, Y.; Fukuyama, R.; Hattori, Y.; Yoshida, C.; Koida, M.; Ogita, K.; Komori, T. RUNX2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J. Cell Biol. 2004, 166, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Gaur, T.; Lengner, C.J.; Hovhannisyan, H.; Bhat, R.A.; Bodine, P.V.; Komm, B.S.; Javed, A.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Canonical Wnt signaling promotes osteogenesis by directly stimulating RUNX2 gene expression. J. Biol. Chem. 2005, 280, 33132–33140. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.F.; Soung do, Y.; Schwarz, E.M.; O’Keefe, R.J.; Drissi, H. Wnt induction of chondrocyte hypertrophy through the RUNX2 transcription factor. J. Cell Physiol. 2006, 208, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Tintut, Y.; Parharni, F.; Li, V.; Karsenty, G.; Demer, L.L. Inhibition of osteoblast-specific transcription factor CBFA1 by the cAMP pathway in osteoblastic cells. Ubiquitin/proteasome-dependent regulation. J. Biol. Chem. 1999, 274, 28875–28879. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, T.M.; Kahler, R.A.; Li, X.; Westendorf, J.J. Histone deacetylase 3 interacts with RUNX2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J. Biol. Chem. 2004, 279, 41998–42007. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.B.; Matsuda, K.; Oh, J.; Barbosa, A.C.; Yang, X.; Meadows, E.; McAnally, J.; Pomajzl, C.; Shelton, J.M.; Richardson, J.A.; et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 2004, 119, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Bensimon, A.; Aebersold, R.; Shiloh, Y. Beyond ATM: The protein kinase landscape of the DNA damage response. FEBS Lett. 2011, 585, 1625–1639. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E.; Jackson, S.P. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011, 25, 409–433. [Google Scholar] [CrossRef] [PubMed]
- Derheimer, F.A.; Kastan, M.B. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett. 2010, 584, 3675–3681. [Google Scholar] [CrossRef] [PubMed]
- Dobbs, T.A.; Tainer, J.A.; Lees-Miller, S.P. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair 2010, 9, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.P.; Siu, W.Y.; Ho, H.T.; Ma, K.H.; Ho, C.C.; Poon, R.Y. Differential contribution of inhibitory phosphorylation of CDC2 and CDK2 for unperturbed cell cycle control and DNA integrity checkpoints. J. Biol. Chem. 2003, 278, 40815–40828. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.P.; Siu, W.Y.; Fung, T.K.; Chan, W.M.; Lau, A.; Arooz, T.; Ng, C.P.; Yamashita, K.; Poon, R.Y. DNA damage during the spindle-assembly checkpoint degrades CDC25A, inhibits cyclin-CDC2 complexes, and reverses cells to interphase. Mol. Biol. Cell 2003, 14, 3989–4002. [Google Scholar] [CrossRef] [PubMed]
- Trovesi, C.; Manfrini, N.; Falcettoni, M.; Longhese, M.P. Regulation of the DNA damage response by cyclin-dependent kinases. J. Mol. Biol. 2013, 425, 4756–4766. [Google Scholar] [CrossRef] [PubMed]
- Wysokinski, D.; Pawlowska, E.; Blasiak, J. RUNX2: A master bone growth regulator that may be involved in the DNA damage response. DNA Cell Biol. 2015, 34, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Bernardin-Fried, F.; Kummalue, T.; Leijen, S.; Collector, M.I.; Ravid, K.; Friedman, A.D. AML1/RUNX1 increases during G1 to S cell cycle progression independent of cytokine-dependent phosphorylation and induces cyclin D3 gene expression. J. Biol. Chem. 2004, 279, 15678–15687. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Shapiro, P.; Fosbrink, M.; Rus, H.; Kumar, R.; Passaniti, A. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by CDC2 regulates endothelial cell proliferation. J. Biol. Chem. 2006, 281, 7118–7128. [Google Scholar] [CrossRef] [PubMed]
- Galindo, M.; Pratap, J.; Young, D.W.; Hovhannisyan, H.; Im, H.J.; Choi, J.Y.; Lian, J.B.; Stein, J.L.; Stein, G.S.; van Wijnen, A.J. The bone-specific expression of RUNX2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J. Biol. Chem. 2005, 280, 20274–20285. [Google Scholar] [CrossRef] [PubMed]
- Zagami, C.J.; Zusso, M.; Stifani, S. RUNX transcription factors: Lineage-specific regulators of neuronal precursor cell proliferation and post-mitotic neuron subtype development. J. Cell. Biochem. 2009, 107, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Rajgopal, A.; Young, D.W.; Mujeeb, K.A.; Stein, J.L.; Lian, J.B.; van Wijnen, A.J.; Stein, G.S. Mitotic control of RUNX2 phosphorylation by both CDK1/cyclin B kinase and PP1/PP2A phosphatase in osteoblastic cells. J. Cell. Biochem. 2007, 100, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Calo, E.; Quintero-Estades, J.A.; Danielian, P.S.; Nedelcu, S.; Berman, S.D.; Lees, J.A. Rb regulates fate choice and lineage commitment in vivo. Nature 2010, 466, 1110–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blyth, K.; Vaillant, F.; Jenkins, A.; McDonald, L.; Pringle, M.A.; Huser, C.; Stein, T.; Neil, J.; Cameron, E.R. RUNX2 in normal tissues and cancer cells: A developing story. Blood Cells Mol. Dis. 2010, 45, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Juric, D.; Lacayo, N.J.; Ramsey, M.C.; Racevskis, J.; Wiernik, P.H.; Rowe, J.M.; Goldstone, A.H.; O’Dwyer, P.J.; Paietta, E.; Sikic, B.I. Differential gene expression patterns and interaction networks in BCR-ABL-positive and -negative adult acute lymphoblastic leukemias. J. Clin. Oncol. 2007, 25, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Heidari, N.; Miller, A.V.; Hicks, M.A.; Marking, C.B.; Harada, H. Glucocorticoid-mediated BIM induction and apoptosis are regulated by RUNX2 and c-Jun in leukemia cells. Cell Death Dis. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Jönsson, S.; Hjorth-Hansen, H.; Olsson, B.; Wadenvik, H.; Sundan, A.; Standal, T. Imatinib inhibits proliferation of human mesenchymal stem cells and promotes early but not late osteoblast differentiation in vitro. J. Bone Miner Metab. 2012, 30, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Pratap, J.; Lian, J.B.; Stein, G.S. Metastatic bone disease: Role of transcription factors and future targets. Bone 2011, 48, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Vladimirova, V.; Waha, A.; Luckerath, K.; Pesheva, P.; Probmeister, R. RUNX2 is expressed in human glioma cells and mediates the expression of galectin-3. J. Neurosci. Res. 2008, 86, 2450–2461. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, T.; Nakagawara, A.; Nagase, H. RUNX family participates in the regulation of p53-dependent DNA damage response. Int. J. Genom. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, T.; Wu, D.; Sugimoto, H.; Nagase, H.; Nakagawara, A. Runt-related transcription factor 2 (RUNX2) inhibits p53-dependent apoptosis through the collaboration with HDAC6 in response to DNA damage. Cell Death Dis. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, C.; Jarzembowski, J.A.; Opipari, A.W., Jr.; Castle, V.P.; Kwok, R.P. HDAC6 deacetylates Ku70 and regulates Ku70-Bax binding in neuroblastoma. Neoplasia 2011, 13, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Corney, D.C.; Flesken-Nikitin, A.; Godwin, A.K.; Wang, W.; Nikitin, A.Y. MicroRNA-34b and microRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007, 67, 8433–8438. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, B. p53 control of bone remodeling. J. Cell. Biochem. 2010, 111, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Lengner, C.J.; Steinman, H.A.; Gagnon, J.; Smith, T.W.; Henderson, J.E.; Kream, B.E.; Stein, G.S.; Lian, J.B.; Jones, S.N. Osteoblast differentiation and skeletal development are regulated by Mdm2-p53 signaling. J. Cell Biol. 2006, 172, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Syed, F.; Khosla, S. Mechanisms of sex steroid effects on bone. Biochem. Biophys. Res. Commun. 2005, 328, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.P.; Boyd, J.; Frank, G.R.; Takahashi, H.; Cohen, R.M.; Specker, B.; Williams, T.C.; Lubahn, D.B.; Korach, K.S. Estrogen resistance caused by a mutation in the estrogen-receptor gene in a man. N. Engl. J. Med. 1994, 331, 1056–1061. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.E.; Boon, W.C.; Proietto, J.; Simpson, E.R. Of mice and men: The evolving phenotype of aromatase deficiency. Trends Endocrinol. Metab. 2006, 17, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Imai, Y.; Matsumoto, T.; Sato, S.; Takeuchi, K.; Igarashi, K.; Harada, Y.; Azuma, Y.; Krust, A.; Yamamoto, Y.; et al. Estrogen prevents bone loss via estrogen receptor α and induction of Fas ligand in osteoclasts. Cell 2007, 130, 811–823. [Google Scholar] [CrossRef] [PubMed]
- Zallone, A. Direct and indirect estrogen actions on osteoblasts and osteoclasts. Ann. N. Y. Acad. Sci. 2006, 1068, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Haji, M.; Nishi, Y.; Yanase, T.; Takayanagi, R.; Nawata, H. Aromatase activity in human osteoblast-like osteosarcoma cell. Calcif. Tissue Int. 1993, 52, 107–109. [Google Scholar] [CrossRef] [PubMed]
- Nawata, H.; Tanaka, S.; Tanaka, S.; Takayanagi, R.; Sakai, Y.; Yanase, T.; Ikuyama, S.; Haji, M. Aromatase in bone cell: Association with osteoporosis in postmenopausal women. J. Steroid Biochem. Mol. Biol. 1995, 53, 165–174. [Google Scholar] [CrossRef]
- Tanaka, S.; Haji, M.; Takayanagi, R.; Tanaka, S.; Sugioka, Y.; Nawata, H. 1,25-dihydroxyvitamin D3 enhances the enzymatic activity and expression of the messenger ribonucleic acid for aromatase cytochrome P450 synergistically with dexamethasone depending on the vitamin D receptor level in cultured human osteoblasts. Endocrinology 1996, 137, 1860–1869. [Google Scholar] [PubMed]
- Jeong, J.H.; Jung, Y.K.; Kim, H.J.; Jin, J.S.; Kim, H.N.; Kang, S.M.; Kim, S.Y.; van Wijnen, A.J.; Stein, J.L.; Lian, J.B.; et al. The gene for aromatase, a rate-limiting enzyme for local estrogen biosynthesis, is a downstream target gene of RUNX2 in skeletal tissues. Mol. Cell. Biol. 2010, 30, 2365–2375. [Google Scholar] [CrossRef] [PubMed]
- Khalid, O.; Baniwal, S.K.; Purcell, D.J.; Leclerc, N.; Gabet, Y.; Stallcup, M.R.; Coetzee, G.A.; Frenkel, B. Modulation of RUNX2 activity by estrogen receptor-α: Implications for osteoporosis and breast cancer. Endocrinology 2008, 149, 5984–5995. [Google Scholar] [CrossRef] [PubMed]
- Kos, M.; Reid, G.; Denger, S.; Gannon, F. Minireview: Genomic organization of the human ERα gene promoter region. Mol. Endocrinol. 2001, 15, 2057–2063. [Google Scholar] [CrossRef] [PubMed]
- Lambertini, E.; Penolazzi, L.; Tavanti, E.; Schincaglia, G.P.; Zennaro, M.; Gambari, R.; Piva, R. Human estrogen receptor α gene is a target of RUNX2 transcription factor in osteoblasts. Exp. Cell Res. 2007, 313, 1548–1560. [Google Scholar] [CrossRef] [PubMed]
- Kammerer, M.; Gutzwiller, S.; Stauffer, D.; Delhon, I.; Seltenmeyer, Y.; Fournier, B. Estrogen receptor α (ERα) and estrogen related receptor α (ERRα) are both transcriptional regulators of the RUNX2-I isoform. Mol. Cell. Endocrinol. 2013, 369, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Galindo, M.; Teplyuk, V.I.; Pratap, J.; Young, D.W.; Lapointe, D.; Javed, A.; Stein, J.L.; Lian, J.B.; Stein, G.S.; et al. RUNX2 regulates G protein-coupled signaling pathways to control growth of osteoblast progenitors. J. Biol. Chem. 2008, 283, 27585–27597. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Zhang, Y.; Lou, Y.; Hawse, J.R.; Hassan, M.Q.; Teplyuk, V.I.; Pratap, J.; Galindo, M.; Stein, J.L.; Stein, G.S.; et al. The osteogenic transcription factor RUNX2 controls genes involved in sterol/steroid metabolism, including CYP11A1 in osteoblasts. Mol. Endocrinol. 2009, 23, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Otsuka, F.; Takano-Narazaki, M.; Katsuyama, T.; Nakamura, E.; Tsukamoto, N.; Inagaki, K.; Sada, K.E.; Makino, H. Estrogen facilitates osteoblast differentiation by upregulating bone morphogenetic protein-4 signaling. Steroids 2013, 78, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Smith, G.H. Murine mammary epithelial stem cells: Discovery, function, and current status. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, N.; McDonald, L.; Morris, J.S.; Cameron, E.R.; Blyth, K. RUNX2 in mammary gland development and breast cancer. J. Cell Physiol. 2013, 228, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Chimge, N.O.; Baniwal, S.K.; Luo, J.; Coetzee, S.; Khalid, O.; Berman, B.P.; Tripathy, D.; Ellis, M.J.; Frenkel, B. Opposing effects of RUNX2 and estradiol on breast cancer cell proliferation: In vitro identification of reciprocally regulated gene signature related to clinical letrozole responsiveness. Clin. Cancer Res. 2012, 18, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Archambault, V.; Glover, D.M. Polo-like kinases: Conservation and divergence in their functions and regulation. Nat. Rev. Mol. Cell Biol. 2009, 10, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Weroha, S.J.; Lingle, W.L.; Papa, D.; Salisbury, J.L.; Li, S.A. Estrogen mediates Aurora-A overexpression, centrosome amplification, chromosomal instability, and breast cancer in female ACI rats. Proc. Natl. Acad. Sci. USA 2004, 101, 18123–18128. [Google Scholar] [CrossRef] [PubMed]
- Javed, A.; Barnes, G.L.; Pratap, J.; Antkowiak, T.; Gerstenfeld, L.C.; van Wijnen, A.J.; Stein, J.L.; Lian, J.B.; Stein, G.S. Impaired intranuclear trafficking of RUNX2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 1454–1459. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Leong, D.T.; Gupta, A.; Shen, L.; Putti, T.; Stein, G.S.; van Wijnen, A.J.; Salto-Tellez, M. Positive association between nuclear RUNX2 and oestrogen-progesterone receptor gene expression characterises a biological subtype of breast cancer. Eur. J. Cancer 2009, 45, 2239–2248. [Google Scholar] [CrossRef] [PubMed]
- Onodera, Y.; Miki, Y.; Suzuki, T.; Takagi, K.; Akahira, J.; Sakyu, T.; Watanabe, M.; Inoue, S.; Ishida, T.; Ohuchi, N.; et al. RUNX2 in human breast carcinoma: Its potential roles in cancer progression. Cancer Sci. 2010, 101, 2670–2675. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.L.; Tarnowski, C.P.; Zhang, J.; Dai, J.; Rohn, E.; Patel, A.H.; Morris, M.D.; Keller, E.T. Bone metastatic LNCaP-derivative C4-2B prostate cancer cell line mineralizes in vitro. Prostate 2001, 47, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Pratap, J.; Javed, A.; Languino, L.R.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. The RUNX2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol. Cell. Biol. 2005, 25, 8581–8591. [Google Scholar] [CrossRef] [PubMed]
- Barnes, G.L.; Javed, A.; Waller, S.M.; Kamal, M.H.; Hebert, K.E.; Hassan, M.Q.; Bellahcene, A.; van Wijnen, A.J.; Young, M.F.; Lian, J.B.; et al. Osteoblast-related transcription factors RUNX2 (CBFA1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res. 2003, 63, 2631–2637. [Google Scholar] [PubMed]
- Zelzer, E.; Glotzer, D.J.; Hartmann, C.; Thomas, D.; Fukai, N.; Soker, S.; Olsen, B.R. Tissue specific regulation of VEGF expression during bone development requires CBFA1/RUNX2. Mech. Dev. 2001, 106, 97–106. [Google Scholar] [CrossRef]
- Taranta, A.; Brama, M.; Teti, A.; de luca, V.; Scandurra, R.; Spera, G.; Agnusdei, D.; Termine, J.D.; Migliaccio, S. The selective estrogen receptor modulator raloxifene regulates osteoclast and osteoblast activity in vitro. Bone 2002, 30, 368–376. [Google Scholar] [CrossRef]
- McCarthy, T.L.; Chang, W.Z.; Liu, Y.; Centrella, M. RUNX2 integrates estrogen activity in osteoblasts. J. Biol. Chem. 2003, 278, 43121–43129. [Google Scholar] [CrossRef] [PubMed]
- Lund, A.H. miR-10 in development and cancer. Cell Death Differ. 2010, 17, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Tehler, D.; Hoyland-Kroghsbo, N.M.; Lund, A.H. The miR-10 microRNA precursor family. RNA Biol. 2011, 8, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, J.; Yang, N.; Greshock, J.; Megraw, M.S.; Giannakakis, A.; Liang, S.; Naylor, T.L.; Barchetti, A.; Ward, M.R.; et al. MicroRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA 2006, 103, 9136–9141. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Luo, Y.C.; Wan, H.Y.; Wang, J.; Zhang, P.P.; Liu, M.; Li, X.; Li, S.; Tang, H. MicroRNA-10a is involved in the metastatic process by regulating Eph tyrosine kinase receptor A4-mediated epithelial-mesenchymal transition and adhesion in hepatoma cells. Hepatology 2013, 57, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Cadoo, K.A.; Fornier, M.N.; Morris, P.G. Biological subtypes of breast cancer: Current concepts and implications for recurrence patterns. Q. J. Nucl. Med. Mol. Imaging 2013, 57, 312–321. [Google Scholar] [PubMed]
- Bouchalova, K.; Cizkova, M.; Cwiertka, K.; Trojanec, R.; Friedecky, D.; Hajduch, M. Lapatinib in breast cancer—The predictive significance of HER1 (EGFR), HER2, PTEN and PIK3CA genes and lapatinib plasma level assessment. Biomed. Pap. 2010, 154, 281–288. [Google Scholar] [CrossRef]
- Skandalis, S.S.; Afratis, N.; Smirlaki, G.; Nikitovic, D.; Theocharis, A.D.; Tzanakakis, G.N.; Karamanos, N.K. Cross-talk between estradiol receptor and EGFR/IGF-IR signaling pathways in estrogen-responsive breast cancers: Focus on the role and impact of proteoglycans. Matrix Biol. 2014, 35, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J. Treatment of HER2-overexpressing breast cancer. Ann. Oncol. 2010, 21 (Suppl. 7), 36–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.J.; Lei, Y.Y.; Mei, J.H.; Wang, C.L. Recent progress in HER2 associated breast cancer. Asian Pac. J. Cancer Prev. 2015, 16, 2591–600. [Google Scholar] [PubMed]
- Nakamura, T.; Toita, H.; Yoshimoto, A.; Nishimura, D.; Takagi, T.; Ogawa, T.; Takeya, T.; Ishida-Kitagawa, N. Potential involvement of Twist2 and Erk in the regulation of osteoblastogenesis by HB-EGF-EGFR signaling. Cell Struct. Funct. 2010, 35, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Barling, P.M.; Lai, A.K.; Nicholson, LF. Distribution of EGF and its receptor in growing red deer antler. Cell Biol. Int. 2005, 29, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Shimizu, E.; Zhang, X.; Partridge, N.C.; Qin, L. EGFR signaling suppresses osteoblast differentiation and inhibits expression of master osteoblastic transcription factors RUNX2 and Osterix. J. Cell. Biochem. 2011, 112, 1749–1760. [Google Scholar] [CrossRef] [PubMed]
- Pratap, J.; Wixted, J.J.; Gaur, T.; Zaidi, S.K.; Dobson, J.; Gokul, K.D.; Hussain, S.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. RUNX2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 2008, 68, 7795–7802. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. Cancer cell cycles. Science 1996, 274, 1672–1677. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.L.; Chen, S.Z.; Chen, W.; Zheng, W.H.; Xia, X.H.; Yang, H.J.; Li, B.; Mao, W.M. The impact of cyclin D1 overexpression on the prognosis of ER-positive breast cancers: A meta-analysis. Breast Cancer Res. Treat. 2013, 139, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.S.; Han, H.S.; Hong, Y.C.; Lee, H.J.; Paik, N.S. Prognostic value of combined analysis of cyclin D1 and estrogen receptor status in breast cancer patients. Pathol. Int. 2003, 53, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Elsheikh, S.; Green, A.R.; Aleskandarany, M.A.; Grainge, M.; Paish, C.E.; Lambros, M.B.; Reis-Filho, J.S.; Ellis, I.O. CCND1 amplification and cyclin D1 expression in breast cancer and their relation with proteomic subgroups and patient outcome. Breast Cancer Res. Treat. 2008, 109, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Aaltonen, K.; Amini, R.M.; Landberg, G.; Eerola, H.; Aittomaki, K.; Heikkila, P.; Nevanlinna, H.; Blomqvist, C. Cyclin D1 expression is associated with poor prognostic features in estrogen receptor positive breast cancer. Breast Cancer Res. Treat. 2009, 113, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Zwijsen, R.M.; Buckle, R.S.; Hijmans, E.M.; Loomans, C.J.; Bernards, R. Ligand-independent recruitment of steroid receptor coactivators to estrogen receptor by cyclin D1. Genes Dev. 1998, 12, 3488–3498. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.Y.; Zelenka, P.S. Cyclins, cyclin-dependent kinases and differentiation. BioEssays 1997, 19, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Casimiro, M.C.; Wang, C.; Li, Z.; Di Sante, G.; Willmart, N.E.; Addya, S.; Chen, L.; Liu, Y.; Lisanti, M.P.; Pestell, R.G. Cyclin D1 determines estrogen signaling in the mammary gland in vivo. Mol. Endocrinol. 2013, 27, 1415–1428. [Google Scholar] [CrossRef] [PubMed]
- Castro-Rivera, E.; Samudio, I.; Safe, S. Estrogen regulation of cyclin D1 gene expression in ZR-75 breast cancer cells involves multiple enhancer elements. J. Biol. Chem. 2001, 276, 30853–30861. [Google Scholar] [CrossRef] [PubMed]
- Owens, T.W.; Rogers, R.L.; Best, S.A.; Ledger, A.; Mooney, A.M.; Ferguson, A.; Shore, P.; Swarbrick, A.; Ormandy, C.J.; Simpson, P.T.; et al. RUNX2 is a novel regulator of mammary epithelial cell fate in development and breast cancer. Cancer Res. 2014, 74, 5277–5286. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Wang, X.; Drissi, H.; Liu, F.; O’Keefe, R.J.; Chen, D. Cyclin D1-Cdk4 induce RUNX2 ubiquitination and degradation. J. Biol. Chem. 2006, 281, 16347–16353. [Google Scholar] [CrossRef] [PubMed]
- Selvamurugan, N.; Kwok, S.; Partridge, N.C. Smad3 interacts with JunB and CBFA1/RUNX2 for transforming growth factor-β1-stimulated collagenase-3 expression in human breast cancer cells. J. Biol. Chem. 2004, 279, 27764–27773. [Google Scholar] [CrossRef] [PubMed]
- Zelzer, E.; McLean, W.; Ng, Y.S.; Reginato, A.M.; Lovejoy, S.; D’Amore, P.A.; Olsen, B.R. Skeletal defects in VEGF120/120 mice reveal multiple roles for VEGF in skeletogenesis. Development 2002, 129, 1893–1904. [Google Scholar] [PubMed]
- Lin, S.Y.; Xia, W.; Wang, J.C.; Kwong, K.Y.; Spohn, B.; Wen, Y.; Pestell, R.G.; Hung, M.C. β-catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. USA 2000, 97, 4262–4266. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, T.M.; Pechenkina, I.V.; Hatsell, S.; Cowin, P. β-catenin and cyclin D1: Connecting development to breast cancer. Cell Cycle 2004, 3, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Zheng, Y.; Li, R.; Li, X.; Zhou, M.; Niu, Y.; Zhang, Q. B-catenin enhances odontoblastic differentiation of dental pulp cells through activation of RUNX2. PLoS ONE 2014, 9, e88890. [Google Scholar] [CrossRef] [PubMed]
- Hua, H.; Li, M.; Luo, T.; Yin, Y.; Jiang, Y. Matrix metalloproteinases in tumorigenesis: An evolving paradigm. Cell Mol. Life Sci. 2011, 68, 3853–3868. [Google Scholar] [CrossRef] [PubMed]
- Przybylowska, K.; Kluczna, A.; Zadrozny, M.; Krawczyk, T.; Kulig, A.; Rykala, J.; Kolacinska, A.; Morawiec, Z.; Drzewoski, J.; Blasiak, J. Polymorphisms of the promoter regions of matrix metalloproteinases genes MMP-1 and MMP-9 in breast cancer. Breast Cancer Res. Treat. 2006, 95, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Yang, J.; Moses, M.A. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J. Clin. Oncol. 2009, 27, 5287–5297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Cao, X.; Liu, Y.; Cao, W.; Zhang, F.; Zhang, S.; Li, H.; Ning, L.; Fu, L.; Niu, Y.; et al. Tumor-derived matrix metalloproteinase-13 (MMP-13) correlates with poor prognoses of invasive breast cancer. BMC Cancer 2008, 83, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Poola, I.; DeWitty, R.L.; Marshalleck, J.J.; Bhatnagar, R.; Abraham, J.; Leffall, L.D. Identification of MMP-1 as a putative breast cancer predictive marker by global gene expression analysis. Nat. Med. 2005, 11, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Têtu, B.; Brisson, J.; Wang, C.S.; Lapointe, H.; Beaudry, G.; Blanchette, C.; Trudel, D. The influence of MMP-14, TIMP-2 and MMP-2 expression on breast cancer prognosis. Breast Cancer Res. 2006, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.M.; Walsh, L.A. Candidate prognostic markers in breast cancer: Focus on extracellular proteases and their inhibitors. Breast Cancer 2014, 6, 81–91. [Google Scholar] [PubMed]
- Achari, Y.; Lu, T.; Hart, D.A. Polymorphisms in the promoter regions for human MMP-1 and MMP-13 lead to differential responses to the α and β isoforms of estrogen receptor and their ligand in vitro. Biochim. Biophys. Acta 2008, 1782, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Achari, Y.; Rattner, J.B.; Hart, D.A. Evidence that estrogen receptor β enhances MMP-13 promoter activity in HIG-82 cells and that this enhancement can be influenced by ligands and involves specific promoter sites. Biochem. Cell Biol. 2007, 85, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Hudelist, G.; Keckstein, J.; Czerwenka, K.; Lass, H.; Walter, I.; Auer, M.; Wieser, F.; Wenzl, R.; Kubista, E.; Singer, C.F. Estrogen receptor β and matrix metalloproteinase 1 are coexpressed in uterine endometrium and endometriotic lesions of patients with endometriosis. Fertil. Steril. 2005, 84, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Ortega, N.; Behonick, D.J.; Werb, Z. Matrix remodeling during endochondral ossification. Trends Cell Biol. 2004, 14, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Selvamurugan, N.; Partridge, N.C. Constitutive expression and regulation of collagenase-3 in human breast cancer cells. Mol. Cell Biol. Res. Commun. 2000, 3, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Xu, X. RUNX2 RNA interference inhibits the invasion of osteosarcoma. Oncol. Lett. 2015, 9, 2455–2458. [Google Scholar] [CrossRef] [PubMed]
- Baniwal, S.K.; Khalid, O.; Gabet, Y.; Shah, R.R.; Purcell, D.J.; Mav, D.; Kohn-Gabet, A.E.; Shi, Y.; Coetzee, G.A.; Frenkel, B. RUNX2 transcriptome of prostate cancer cells: Insights into invasiveness and bone metastasis. Mol. Cancer 2010, 9. [Google Scholar] [CrossRef] [PubMed]
- Kayed, H.; Jiang, X.; Keleg, S.; Jesnowski, R.; Giese, T.; Berger, M.R.; Esposito, I.; Löhr, M.; Friess, H.; Kleeff, J. Regulation and functional role of the runt-related transcription factor-2 in pancreatic cancer. Br. J. Cancer 2007, 97, 1106–1115. [Google Scholar] [CrossRef] [PubMed]
- Hayami, T.; Kapila, Y.L.; Kapila, S. MMP-1 (collagenase-1) and MMP-13 (collagenase-3) differentially regulate markers of osteoblastic differentiation in osteogenic cells. Matrix Biol. 2008, 27, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Al-Azzawi, F.; Wahab, M. Estrogen and colon cancer: Current issues. Climacteric 2002, 5, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.L.; Ballard-Barbash, R.; Edwards, S.; Caan, B.J.; Potter, J.D. Body mass index and colon cancer: An evaluation of the modifying effects of estrogen (United States). Cancer Causes Control. 2003, 14, 75–84. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wysokinski, D.; Blasiak, J.; Pawlowska, E. Role of RUNX2 in Breast Carcinogenesis. Int. J. Mol. Sci. 2015, 16, 20969-20993. https://doi.org/10.3390/ijms160920969
Wysokinski D, Blasiak J, Pawlowska E. Role of RUNX2 in Breast Carcinogenesis. International Journal of Molecular Sciences. 2015; 16(9):20969-20993. https://doi.org/10.3390/ijms160920969
Chicago/Turabian StyleWysokinski, Daniel, Janusz Blasiak, and Elzbieta Pawlowska. 2015. "Role of RUNX2 in Breast Carcinogenesis" International Journal of Molecular Sciences 16, no. 9: 20969-20993. https://doi.org/10.3390/ijms160920969