Duplex High-Resolution Melting Assay for the Simultaneous Genotyping of IL28B rs12979860 and PNPLA3 rs738409 Polymorphisms in Chronic Hepatitis C Patients
Abstract
:1. Introduction
2. Results
2.1. Primer Characterization in Singleplex Reactions
| Parameter | Value |
|---|---|
| Age, Mean (SD) | 52 (10) |
| Gender: n (%) | |
| male | 58 (44) |
| female | 74 (56) |
| BMI, mean (SD) | 25.7 (4.1) |
| Fibrosis staging: n (%) | |
| F0–F2 | 64 (54.2) |
| F3–F4 | 54 (45.8) |
| Activity grading: n (%) | |
| A0–A1 | 28 (23.7) |
| A2–A3 | 90 (76.3) |
| HCV RNA × 103 IU/mL, median (Q25–Q75) | 723 (197–2161) |
| Sustained viral response (SVR), n (%) | |
| positive | 44 (42.7) |
| negative | 59 (57.3) |
| Genotype IL28B rs12979860, n (%) | |
| CC | 35 (26.5) |
| CT | 74 (56.1) |
| TT | 23 (17.4) |
| minor allele frequency | 0.4545 |
| Genotype PNPLA3 rs738409, n (%) | |
| CC | 69 (52.3) |
| CG | 58 (43.9) |
| GG | 5 (3.8) |
| minor allele frequency | 0.2576 |
| Diplotype No. | rs12979860 | rs738409 | No. (%) of Cases with Shown Diplotype |
|---|---|---|---|
| 1 | CC | CC | 22 (16.7) |
| 2 | CC | CG | 13 (9.8) |
| 3 | CC | GG | 0 |
| 4 | CT | CC | 39 (29.5) |
| 5 | CT | CG | 31 (23.5) |
| 6 | CT | GG | 4 (3.0) |
| 7 | TT | CC | 8 (6.1) |
| 8 | TT | CG | 14 (10.6) |
| 9 | TT | GG | 1 (0.8) |
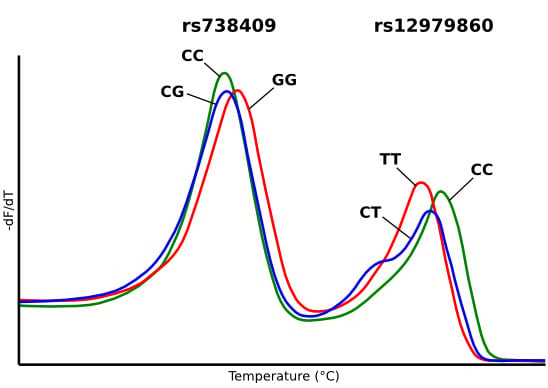
2.2. Characterization of Duplex HRM Assay



2.3. Discussions

3. Experimental Section
3.1. Patients and Samples
3.2. Primer Design
3.3. Construction of Melting Standards
3.4. Real-Time PCR and High-Resolution Melting Analysis
| Primer Name a | Nucleotide Sequence (5′→3′) b | Amplicon Length (bp) | Corresponding Human Genomic Region c |
|---|---|---|---|
| rs12979860HRMf | CGAGGCGACCACGGAGCTCCCCGAAGGC | - | 39248130–39248145 |
| rs12979860HRMr | GAGCGCGGAGTGCAATTC | 59 | 39248176–39248159 |
| rs12979860aspf | TCTGCACAGTCTGGGATTCC | - | 39248055–39248074 |
| rs12979860aspCr | CGGCGGGGCGGCCGAGTGCAATTCAACCCTGGTTGG | 128 | 39248169–39248147 |
| rs12979860aspTr | GAGTGCAATTCAACCCTGGTTGA | 115 | 39248169–39248147 |
| rs12979860Seqf | CATAGCATTTTTATCCACCTCTGCACAGTCTGGGAT | - | 39248052–39248071 |
| rs12979860Seqr | CAAATTGTGAATTCGCGCGGAGTGCAATTCAAC | 153 | 39248174–39248156 |
| rs12979860cf | GGACGAGAGGGCGTTAGAG | - | 39247858–39247876 |
| rs12979860cr | GTGCACGGTGATCGCAGAAG | 675 | 39248532–39248513 |
| rs12979860mCr | CAACCCTGGTTCGCGCCTTC | 302 | 39248159–39248140 |
| rs12979860mTr | CAATTCAACCCTGGTTCACGCCTTC | 307 | 39248164–39248140 |
| rs12979860mCf | GAAGGCGCGAACCAGGGTTG | 393 | 39248140–39248159 |
| rs12979860mTf | GAAGGCGTGAACCAGGGTTGAATTG | 393 | 39248140–39248164 |
| rs738409HRMf | GCCTTGGTATGTTCCTGCTTC | - | 43928824–43928844 |
| rs738409HRMr | GGATAAGGCCACTGTAGAAGG | 46 | 43928869–43928849 |
| rs738409aspGf | CGCGGCGGCCCCTTGGTATGTTCCTGCTTCACG | 97 | 43928825–43928847 |
| rs738409aspCf | CCTTGGTATGTTCCTGCTTCAAC | 87 | 43928825–43928847 |
| rs738409aspr | CTAGCAGAGAAAGCCGACTTAC | - | 43928911–43928890 |
| rs738409Seqf | ACTGAGCGAATTCTTGCTTTCACAGGCCTTGG | - | 43928812–43928830 |
| rs738409Seqr | CATGCTGGAATTCCGCTAGCAGAGAAAGCCGAC | 128 | 43928913–43928894 |
| rs738409cf | CACCGATCTAGCCCCTTTCA | - | 43928498–43928517 |
| rs738409cr | TAAGTTTTGCTGCCCGGGTA | 583 | 43929080–43929061 |
| rs738409mGr | CTGTAGAAGGGCATGAAGCAGGAAC | 361 | 43928858–43928834 |
| rs738409mCr | CTGTAGAAGGGGATGAAGCAGGAAC | 361 | 43928858–43928834 |
| rs738409mGf | GTTCCTGCTTCATGCCCTTCTACAG | 247 | 43928834–43928858 |
| rs738409mCf | GTTCCTGCTTCATCCCCTTCTACAG | 247 | 43928834–43928858 |
3.5. Reproducibility
3.6. Sanger Sequencing
3.7. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gower, E.; Estes, C.; Blach, S.; Razavi-Shearer, K.; Razavi, H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J. Hepatol. 2014, 61, S45–S57. [Google Scholar] [CrossRef] [PubMed]
- Micallef, J.M.; Kaldor, J.M.; Dore, G.J. Spontaneous viral clearance following acute hepatitis C infection: A systematic review of longitudinal studies. J. Viral Hepat. 2006, 13, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Westbrook, R.H.; Dusheiko, G. Natural history of hepatitis C. J. Hepatol. 2014, 61, S58–S68. [Google Scholar] [CrossRef] [PubMed]
- Bugianesi, E.; Salamone, F.; Negro, F. The interaction of metabolic factors with HCV infection: Does it matter? J. Hepatol. 2012, 56 (Suppl. 1), S56–S65. [Google Scholar] [CrossRef]
- Leandro, G.; Mangia, A.; Hui, J.; Fabris, P.; Rubbia–Brandt, L.; Colloredo, G.; Adinolfi, L.E.; Asselah, T.; Jonsson, J.R.; Smedile, A.; et al. Relationship between steatosis, inflammation, and fibrosis in chronic Hepatitis C: A Meta-Analysis of Individual Patient Data. Gastroenterology 2006, 130, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Fried, M.W.; Shiffman, M.L.; Reddy, K.R.; Smith, C.; Marinos, G.; Gonçales, F.L.; Häussinger, D.; Diago, M.; Carosi, G.; Dhumeaux, D.; et al. Peginterferon Alfa-2a plus Ribavirin for Chronic Hepatitis C Virus Infection. N. Engl. J. Med. 2002, 347, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, S. Antiviral therapies for chronic hepatitis C virus infection with cirrhosis. World J. Hepatol. 2015, 7, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Ford, N.; Swan, T.; Beyer, P.; Hirnschall, G.; Easterbrook, P.; Wiktor, S. Simplification of antiviral hepatitis C virus therapy to support expanded access in resource-limited settings. J. Hepatol. 2014, 61, S132–S138. [Google Scholar] [CrossRef] [PubMed]
- Susser, S.; Herrmann, E.; Lange, C.; Hamdi, N.; Müller, T.; Berg, T.; Perner, D.; Zeuzem, S.; Sarrazin, C. Predictive Value of Interferon-Lambda Gene Polymorphisms for Treatment Response in Chronic Hepatitis C. PLoS ONE 2014, 9, e112592. [Google Scholar] [CrossRef] [PubMed]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Dufour, J.-F.; Muellhaupt, B.; Gerlach, T.; Heim, M.; Moradpour, D.; Cerny, A.; Malinverni, R.; Kaddai, V.; Bochud, M.; et al. Viral genotype-specific role of PNPLA3, PPARG, MTTP, and IL28B in hepatitis C virus-associated steatosis. J. Hepatol. 2011, 55, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Trépo, E.; Pradat, P.; Potthoff, A.; Momozawa, Y.; Quertinmont, E.; Gustot, T.; Lemmers, A.; Berthillon, P.; Amininejad, L.; Chevallier, M.; et al. Impact of patatin-like phospholipase-3 (rs738409 C>G) polymorphism on fibrosis progression and steatosis in chronic hepatitis C. Hepatology 2011, 54, 60–69. [Google Scholar]
- Valenti, L.; Rumi, M.; Galmozzi, E.; Aghemo, A.; del Menico, B.; de Nicola, S.; Dongiovanni, P.; Maggioni, M.; Fracanzani, A.L.; Rametta, R.; et al. Patatin-Like phospholipase domain-containing 3 I148M polymorphism, steatosis, and liver damage in chronic hepatitis C. Hepatology 2011, 53, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Yasui, K.; Kawaguchi, T.; Shima, T.; Mitsuyoshi, H.; Seki, K.; Sendo, R.; Mizuno, M.; Itoh, Y.; Matsuda, F.; Okanoue, T. Effect of PNPLA3 rs738409 variant (I148 M) on hepatic steatosis, necroinflammation, and fibrosis in Japanese patients with chronic hepatitis C. J. Gastroenterol. 2015, 50, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, M.; Grünhage, F.; Lammert, F. Identification of Combined Genetic Determinants of Liver Stiffness within the SREBP1c-PNPLA3 Pathway. Int. J. Mol. Sci. 2013, 14, 21153–21166. [Google Scholar] [CrossRef] [PubMed]
- Valenti, L.; Aghemo, A.; Stättermayer, A.F.; Maggioni, P.; de Nicola, S.; Motta, B.M.; Rumi, M.G.; Dongiovanni, P.; Ferenci, P.; Colombo, M.; et al. Implications of PNPLA3 polymorphism in chronic hepatitis C patients receiving peginterferon plus ribavirin. Aliment. Pharmacol. Ther. 2012, 35, 1434–1442. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Tao, A.; Zhang, S.; Deng, Y.; Chen, G. Association Between Patatin-Like Phospholipase Domain Containing 3 Gene (PNPLA3) Polymorphisms and Nonalcoholic Fatty Liver Disease: A HuGE Review and Meta-Analysis. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.-H.; Li, Y.-L.; Li, D.; Wang, N.-N.; Jing, L.; Huang, Y.-H. The rs738409 (I148M) variant of thePNPLA3 gene and cirrhosis: A meta-analysis. J. Lipid Res. 2015, 56, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Manjunath, H.; Yopp, A.C.; Beg, M.S.; Marrero, J.A.; Gopal, P.; Waljee, A.K. The Effect of PNPLA3 on Fibrosis Progression and Development of Hepatocellular Carcinoma: A Meta-analysis. Am. J. Gastroenterol. 2014, 109, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Trépo, E.; Nahon, P.; Bontempi, G.; Valenti, L.; Falleti, E.; Nischalke, H.-D.; Hamza, S.; Corradini, S.G.; Burza, M.A.; Guyot, E.; et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology 2014, 59, 2170–2177. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Nishida, N.; Sugiyama, M.; Kurosaki, M.; Matsuura, K.; Sakamoto, N.; Nakagawa, M.; Korenaga, M.; Hino, K.; Hige, S.; et al. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat. Genet. 2009, 41, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Suppiah, V.; Moldovan, M.; Ahlenstiel, G.; Berg, T.; Weltman, M.; Abate, M.L.; Bassendine, M.; Spengler, U.; Dore, G.J.; Powell, E.; et al. IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nat. Genet. 2009, 41, 1100–1104. [Google Scholar] [CrossRef] [PubMed]
- Ge, D.; Fellay, J.; Thompson, A.J.; Simon, J.S.; Shianna, K.V.; Urban, T.J.; Heinzen, E.L.; Qiu, P.; Bertelsen, A.H.; Muir, A.J.; et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009, 461, 399–401. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Sousa, M.A.; Fernández-Rodríguez, A.; Guzmán-Fulgencio, M.; García-Álvarez, M.; Resino, S. Meta-analysis: Implications of interleukin-28B polymorphisms in spontaneous and treatment-related clearance for patients with hepatitis C. BMC Med. 2013, 116. [Google Scholar] [CrossRef] [PubMed]
- Chayama, K.; Hayes, C.N.; Abe, H.; Miki, D.; Ochi, H.; Karino, Y.; Toyota, J.; Nakamura, Y.; Kamatani, N.; Sezaki, H.; et al. IL28B But Not ITPA Polymorphism Is Predictive of Response to Pegylated Interferon, Ribavirin, and Telaprevir Triple Therapy in Patients With Genotype 1 Hepatitis C. J. Infect. Dis. 2011, 204, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Bota, S.; Sporea, I.; Şirli, R.; Neghină, A.M.; Popescu, A.; Străin, M. Role of Interleukin-28B Polymorphism as a Predictor of Sustained Virological Response in Patients with Chronic Hepatitis C Treated with Triple Therapy: A Systematic Review and Meta-Analysis. Clin. Drug Investig. 2013, 33, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Hashem, A.M.; Leung, R.; Romero-Gomez, M.; Berg, T.; Dore, G.J.; Chan, H.L.K.; Irving, W.L.; Sheridan, D.; Abate, M.L.; et al. International Hepatitis C Genetics Consortium (IHCGC) Interferon-λ rs12979860 genotype and liver fibrosis in viral and non-viral chronic liver disease. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Kondo, M.; Tateishi, R.; Fujiwara, N.; Kato, N.; Yoshida, H.; Taguri, M.; Koike, K. Impact of IL28B Genetic Variation on HCV-Induced Liver Fibrosis, Inflammation, and Steatosis: A Meta-Analysis. PLoS ONE 2014, 9, e91822. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosio, R.; Aghemo, A.; de Francesco, R.; Rumi, M.G.; Galmozzi, E.; de Nicola, S.; Cheroni, C.; Clark, P.J.; Ronchi, G.; Lampertico, P.; et al. The Association of Il28b Genotype with the Histological Features of Chronic Hepatitis C Is HCV Genotype Dependent. Int. J. Mol. Sci. 2014, 15, 7213–7224. [Google Scholar]
- EASL Recommendations on Treatment of Hepatitis C. Available online: http://files.easl.eu/easl-recommendations-on-treatment-of-hepatitis-C/index.html (accessed on 29 March 2015).
- Fischer, J.; Böhm, S.; Scholz, M.; Müller, T.; Witt, H.; George, J.; Sarrazin, C.; Susser, S.; Schott, E.; Suppiah, V.; et al. Combined effects of different interleukin-28B gene variants on the outcome of dual combination therapy in chronic hepatitis C virus type 1 infection. Hepatology 2012, 55, 1700–1710. [Google Scholar] [CrossRef] [PubMed]
- Van der Stoep, N.; van Paridon, C.D.M.; Janssens, T.; Krenkova, P.; Stambergova, A.; Macek, M.; Matthijs, G.; Bakker, E. Diagnostic guidelines for high-resolution melting curve (HRM) analysis: An interlaboratory validation of BRCA1 mutation scanning using the 96-well LightScanner™. Hum. Mutat. 2009, 30, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, M.G.; Durtschi, J.D.; Wittwer, C.T.; Voelkerding, K.V. Expanded Instrument Comparison of Amplicon DNA Melting Analysis for Mutation Scanning and Genotyping. Clin. Chem. 2007, 53, 1544–1548. [Google Scholar] [CrossRef] [PubMed]
- Rüeger, S.; Bochud, P.-Y.; Dufour, J.-F.; Müllhaupt, B.; Semela, D.; Heim, M.H.; Moradpour, D.; Cerny, A.; Malinverni, R.; Booth, D.R.; et al. Impact of common risk factors of fibrosis progression in chronic hepatitis C. Gut 2015, 64, 1605–1615. [Google Scholar] [CrossRef] [PubMed]
- Ampuero, J.; del Campo, J.A.; Rojas, L.; García-Lozano, J.R.; Solá, R.; Andrade, R.; Pons, J.A.; Navarro, J.M.; Calleja, J.L.; Buti, M.; et al. PNPLA3 rs738409 causes steatosis according to viral & IL28B genotypes in hepatitis C. Ann. Hepatol. 2014, 13, 356–363. [Google Scholar] [PubMed]
- Valenti, L.; Aghemo, A.; Stättermayer, A.F. Interaction between IL28B and PNPLA3 genotypes in the pathogenesis of steatosis in chronic hepatitis C non genotype-3 patients. J. Hepatol. 2012, 56, 1209–1210. [Google Scholar] [CrossRef] [PubMed]
- King, L.Y.; Johnson, K.B.; Zheng, H.; Wei, L.; Gudewicz, T.; Hoshida, Y.; Corey, K.E.; Ajayi, T.; Ufere, N.; Baumert, T.F.; et al. Host Genetics Predict Clinical Deterioration in HCV-Related Cirrhosis. PLoS ONE 2014, 9, e114747. [Google Scholar] [CrossRef] [PubMed]
- Conteduca, V.; Sansonno, D.; Russi, S.; Pavone, F.; Dammacco, F. Therapy of chronic hepatitis C virus infection in the era of direct-acting and host-targeting antiviral agents. J. Infect. 2014, 68, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Hoofnagle, J.H.; Sherker, A.H. Therapy for Hepatitis C—The Costs of Success. N. Engl. J. Med. 2014, 370, 1552–1553. [Google Scholar] [CrossRef] [PubMed]
- Strazzulla, A.; Costa, C.; Pisani, V.; De Maria, V.; Giancotti, F.; Di Salvo, S.; Parisi, S.; Basso, M.; Franzetti, M.; Marascio, N.; et al. Present, old and future strategies for anti-HCV treatment in patients infected by genotype-1: estimation of the drug costs in the Calabria Region in the era of the directly acting antivirals. BMC Infect. Dis. 2014, 14 (Suppl. 5). [Google Scholar] [CrossRef] [PubMed]
- Brogan, A.J.; Talbird, S.E.; Thompson, J.R.; Miller, J.D.; Rubin, J.; Deniz, B. Cost-effectiveness of Telaprevir Combination Therapy for Chronic Hepatitis C. PLoS ONE 2014, 9, e90295. [Google Scholar] [CrossRef] [PubMed]
- Petta, S.; Cabibbo, G.; Enea, M.; Macaluso, F.S.; Plaia, A.; Bruno, R.; Gasbarrini, A.; Bruno, S.; Craxì, A.; Cammà, C. Personalized cost-effectiveness of boceprevir-based triple therapy for untreated patients with genotype 1 chronic hepatitis C. Dig. Liver Dis. 2014, 46, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Pho, M.T.; Linas, B.P. Valuing cure: Bridging cost-effectiveness and coverage decisions for hepatitis C therapy. Hepatology 2014, 60, 12–14. [Google Scholar] [CrossRef] [PubMed]
- Stättermayer, A.F.; Scherzer, T.; Beinhardt, S.; Rutter, K.; Hofer, H.; Ferenci, P. Review article: Genetic factors that modify the outcome of viral hepatitis. Aliment. Pharmacol. Ther. 2014, 39, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Pho, M.T.; Jensen, D.M.; Meltzer, D.O.; Kim, A.Y.; Linas, B.P. Clinical impact of treatment timing for chronic hepatitis C infection: a decision model. J. Viral Hepat. 2015, 22, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.W.S.; Holt, D.C.; Andersson, P.; Giffard, P.M. DNA Concentration Can Specify DNA Melting Point in a High-Resolution Melting Analysis Master Mix. Clin. Chem. 2014, 60, 414–416. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Higami, K.; Masaki, N.; Sugiyama, M.; Mukaide, M.; Saito, H.; Aoki, Y.; Sato, Y.; Imamura, M.; Murata, K.; et al. The rs8099917 Polymorphism, WHEN determined by a Suitable genotyping method, is a better predictor for response to pegylated alpha interferon/ribavirin therapy in Japanese Patients than Other Single Nucleotide Polymorphisms Associated with Interleukin-28B. J. Clin. Microbiol. 2011, 49, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Lefever, S.; Pattyn, F.; Hellemans, J.; Vandesompele, J. Single-Nucleotide Polymorphisms and Other Mismatches Reduce Performance of Quantitative PCR Assays. Clin. Chem. 2013, 59, 1470–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Zhou, L.; Palais, R.A.; Wittwer, C.T. Genotyping Accuracy of High-Resolution DNA Melting Instruments. Clin. Chem. 2014, 60, 864–872. [Google Scholar] [CrossRef] [PubMed]
- uDesign—Web tool for small amplicon design and SNV genotyping. Available online: https://www.dna.utah.edu/udesign/index.html (accessed on 28 February 2013).
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- OligoAnalyzer 3.1. Available online: http://eu.idtdna.com/calc/analyzer (accessed on 28 February 2013).
- Rodriguez, S.; Gaunt, T.R.; Day, I.N.M. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am. J. Epidemiol. 2009, 169, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Gaunt, T.R.; Rodríguez, S.; Day, I.N. Cubic exact solutions for the estimation of pairwise haplotype frequencies: implications for linkage disequilibrium analyses and a web tool “CubeX.”. BMC Bioinform. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- OEGE-Online Encyclopedia for Genetic Epidemiology studies. Available online: http://oege.org/software/ (accessed on 9 February 2015).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enache, E.L.; Sin, A.; Bancu, L.; Ramière, C.; Diaz, O.; André, P.; Enache, L.S. Duplex High-Resolution Melting Assay for the Simultaneous Genotyping of IL28B rs12979860 and PNPLA3 rs738409 Polymorphisms in Chronic Hepatitis C Patients. Int. J. Mol. Sci. 2015, 16, 22223-22242. https://doi.org/10.3390/ijms160922223
Enache EL, Sin A, Bancu L, Ramière C, Diaz O, André P, Enache LS. Duplex High-Resolution Melting Assay for the Simultaneous Genotyping of IL28B rs12979860 and PNPLA3 rs738409 Polymorphisms in Chronic Hepatitis C Patients. International Journal of Molecular Sciences. 2015; 16(9):22223-22242. https://doi.org/10.3390/ijms160922223
Chicago/Turabian StyleEnache, Elena L., Anca Sin, Ligia Bancu, Christophe Ramière, Olivier Diaz, Patrice André, and Liviu S. Enache. 2015. "Duplex High-Resolution Melting Assay for the Simultaneous Genotyping of IL28B rs12979860 and PNPLA3 rs738409 Polymorphisms in Chronic Hepatitis C Patients" International Journal of Molecular Sciences 16, no. 9: 22223-22242. https://doi.org/10.3390/ijms160922223