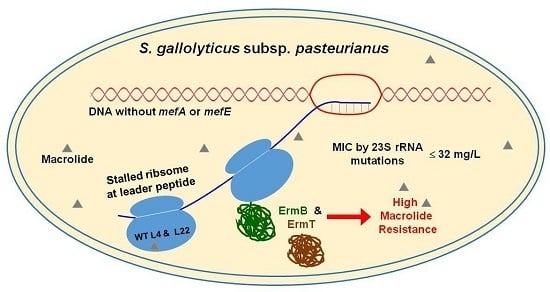
Inducible Expression of both ermB and ermT Conferred High Macrolide Resistance in Streptococcus gallolyticus subsp. pasteurianus Isolates in China
Abstract
:1. Introduction
2. Results
2.1. All S. gallolyticus subsp. pasteurianus Isolates Were Multi-Drug Resistant, with High Level Macrolide Resistance
2.2. The High Macrolide Resistance Was neither Mainly Caused by Efflux Pump nor by Mutations within L4, L22, or 23S rRNA
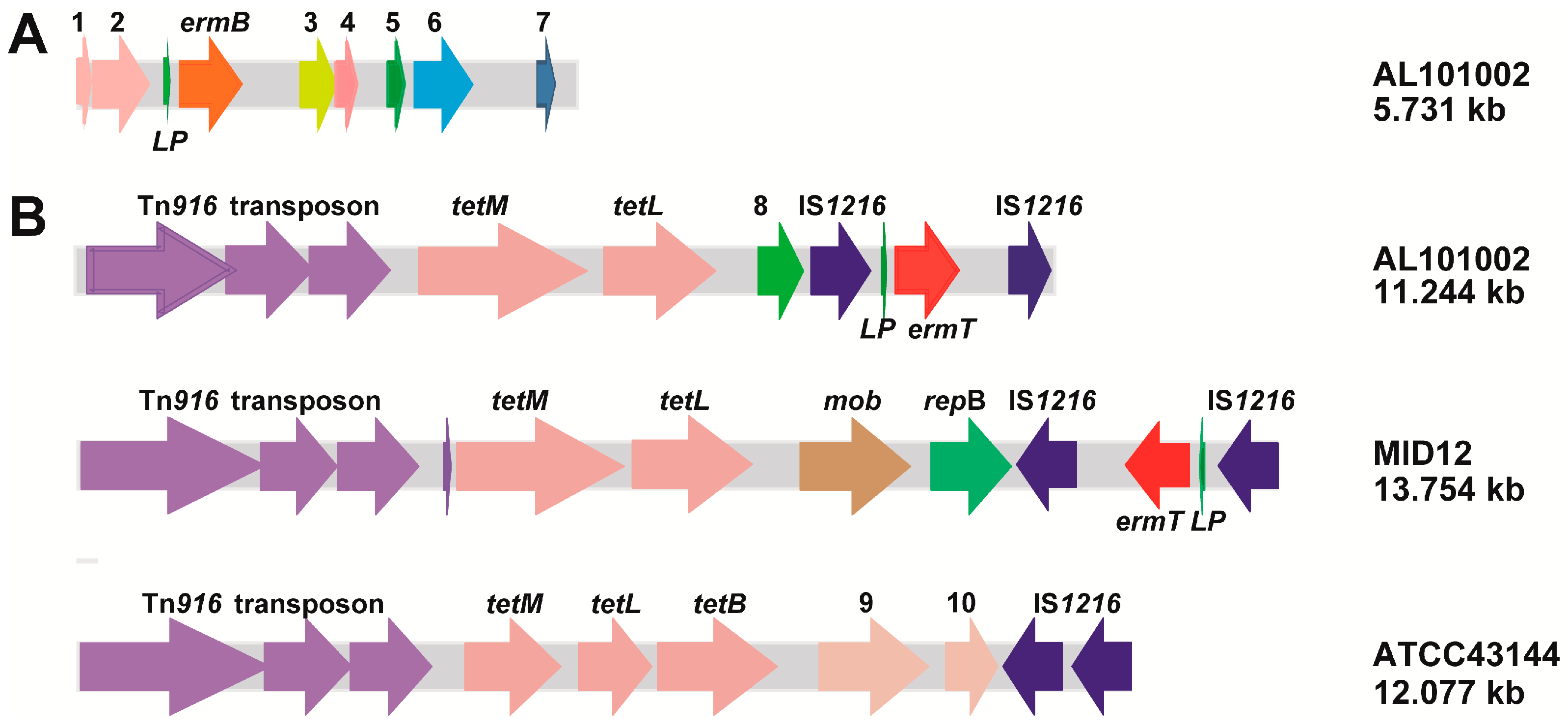
2.3. The ermB and ermT Genes Were Integrated into All Isolate Genomes
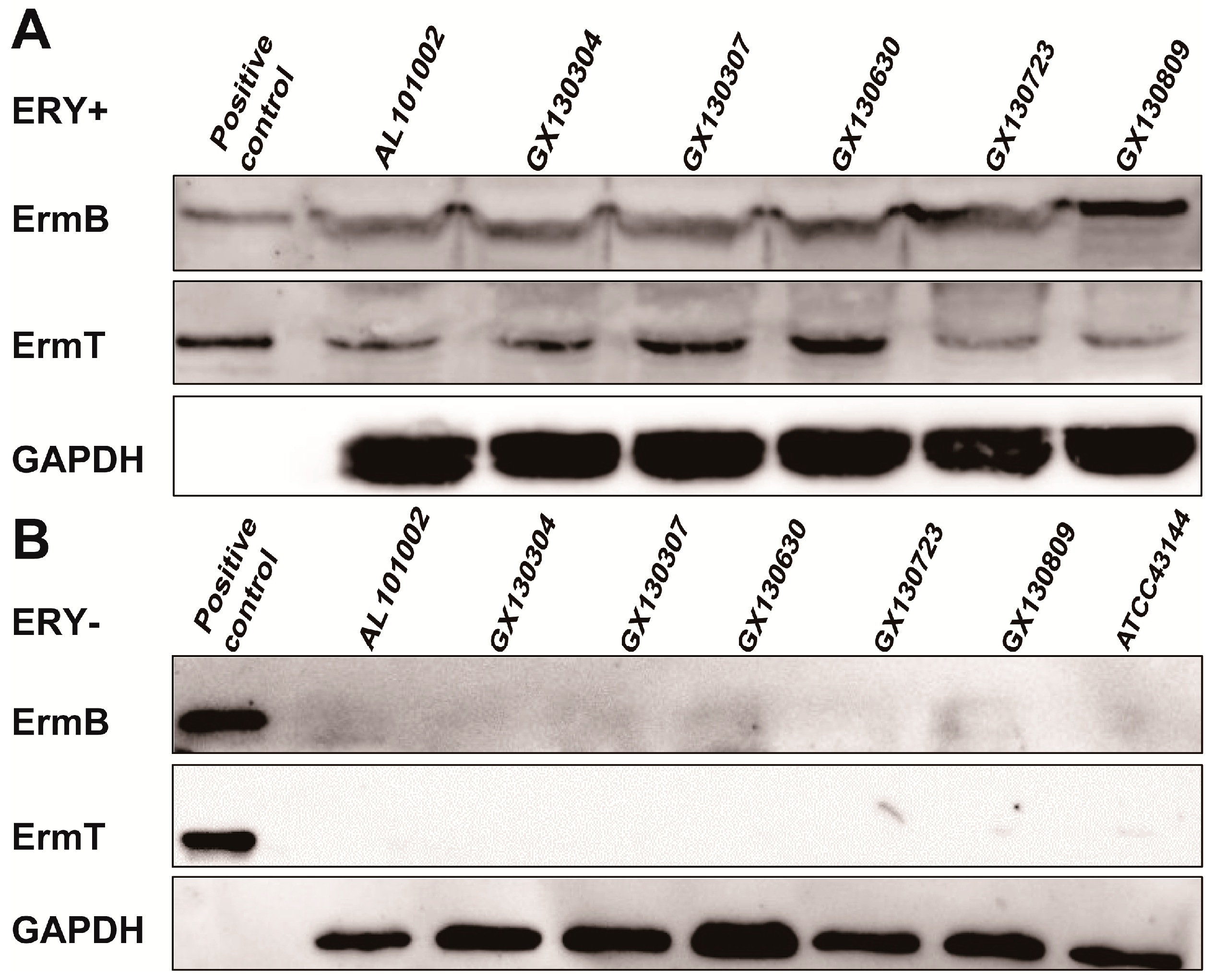
2.4. Both ermB and ermT Were Expressed in the Isolates, and the Expression Was Erythromycin-Inducible
2.5. The Inducible Expression of ermB and ermT Conferred High Level of Macrolide Resistance
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates and Culture Conditions
4.2. Antimicrobial Susceptibility Testing
4.3. Sequence Analysis and Molecular Cloning
4.4. Whole-Genome Sequencing
4.5. Western Blot Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MIC | Minimum inhibitory concentration |
| ATCC | American type culture collection |
| CLSI | Clinical and laboratory standards institute |
| MLS | Macrolide-lincosamide-streptogramin B |
| TSB | Trypticase soy broth |
| MH | Mueller-hinton |
| IPTG | Isopropyl β-d-1-thiogalactopyranoside |
| ECL | Enhanced chemiluminescence |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| PAβN | Phenylalanine-arginine beta-naphthylamide |
References
- Osawa, R.; Fujisawa, T.; Sly, L.I. Streptococcus gallolyticus sp. nov.; gallate degrading organisms formerly assigned to Streptococcus bovis. Syst. Appl. Microbiol. 1995, 18, 1942–1946. [Google Scholar] [CrossRef]
- Ruoff, K.L.; Whiley, R.A.; Beighton, D. Streptococcus. In Manual of Clinical Microbiology, 8th ed.; ASM Press: Washington, DC, USA, 2003; pp. 405–421. [Google Scholar]
- Poyart, C.; Quesne, G.; Trieu-Cuot, P. Taxonomic dissection of the Streptococcus bovis group by analysis of manganese-dependent superoxide dismutase gene (sodA) sequences: Reclassification of ′Streptococcus infantarius subsp. coli′ as Streptococcus lutetiensis sp. nov. and of Streptococcus bovis biotype 11.2 as Streptococcus pasteurianus sp. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 1247–1255. [Google Scholar] [PubMed]
- Schlegel, L.; Grimont, F.; Ageron, E.; Grimont, P.A.D.; Bouvet, A. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: Description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 632–645. [Google Scholar]
- Li, M.; Gu, C.; Zhang, W.; Li, S.; Liu, J.; Qin, C.; Su, J.; Cheng, G.; Hu, X. Isolation and characterization of Streptococcus gallolyticus subsp. pasteurianus causing meningitis in ducklings. Vet. Microbiol. 2013, 162, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Kimpe, A.; Decostere, A.; Martel, A.; Pevriese, L.A.; Haesebrouck, F. Phenotypic and genetic characterization of resistance against macrolides and lincosamides in Streptococcus gallolyticus strains isolated from pigeons and humans. Microb. Drug Resist. 2003, 9, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J.; Ainsworth, H.; Boon, J.D.; Twomey, D.F. Streptococcus gallolyticus subsp. pasteurianus septicaemia in goslings. Vet. J. 2008, 176, 251–253. [Google Scholar] [PubMed]
- Saumya, D.; Wietunge, S.; Dunn, P.; Wallner-Pendleton, E.; Lintner, V.; Matthews, T.; Pierre, T.; Kariyawasam, S. Acute septicemia caused by Streptococcus gallolyticus subsp. pasteurianus in turkey poults. Avian Dis. 2014, 58, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Klatte, J.M.; Clarridge, J.E.; Bratcher, D.; Selvarangan, R. A longitudinal case series description of meningitis due to Streptococcus gallolyticus subsp. pasteurianus in infants. J. Clin. Microbiol. 2012, 50, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Nagamatsu, M.; Takaqi, T.; Ohyanagi, T.; Yamazaki, S.; Nobuoka, S.; Takemura, H.; Akita, H.; Miyai, M.; Ohkusu, K. Neonatal meningitis caused by Streptococcus gallolyticus subsp. pasteurianus. J. Infect. Chemother. 2012, 18, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Ishiwada, N.; Tanaka, J.; Okusu, K.; Ichimura, S.; Hishiki, H.; Ota, S.; Kohno, Y. Streptococcus gallolyticus subsp. pasteurianus meningitis in an infant. Pediatr. Int. 2014, 56, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Hede, S.V. Diagnosis and treatment of childhood meningitis caused by Streptococcus bovis group. Curr. Infect. Dis. Rep. 2016, 18, 11. [Google Scholar] [CrossRef] [PubMed]
- Marmonlin, E.S.; Hartmeyer, G.N.; Christensen, J.J.; Nielsen, X.C.; Dargis, R.; Skov, M.N.; Knudsen, E.; Kemp, M.; Justesen, U.S. Bacteremia with the bovis group streptococci: Species identification and association with infective endocarditis and with gastrointestinal disease. Diagn. Microbiol. Infect. Dis. 2016, 85, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Sturt, A.S.; Yang, L.; Sandhu, K.; Pei, Z.H.; Cassai, N.; Blaser, M.J. Streptococcus gallolyticus subspecies pasteurianus (biotype II/2), a newly reported cause of adult meningitis. J. Clin. Microbiol. 2010, 48, 2247–2249. [Google Scholar] [CrossRef] [PubMed]
- Lazarovitch, T.; Sango, M.; Levine, M.; Brusovansky, R.; Akins, R.; Hayakawa, K.; Lephart, P.R.; Sobel, J.D.; Kaye, K.S.; Marchaim, D. The relationship between the new taxonomy of Streptococcus bovis and its clonality to colon cancer, endocarditis, and biliary disease. Infection 2013, 41, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Takamura, N.; Kenza, T.; Minami, K.; Matsumura, M. Infective endocarditis caused by Streptococcus gallolyticus subspecies pasteurianus and colon cancer. BMJ Case Rep. 2014. [Google Scholar] [CrossRef] [PubMed]
- Chirouze, C.; Patry, I.; Duval, X.; Baty, V.; Tattevin, P.; Aparicio, T.; Pagenault, M.; Carbonnel, F.; Couetdic, G.; Hoen, B. Streptococcus bovis/Streptococcus equinus complex fecal carriage, colorectal carcinoma, and infective endocarditis: A new appraisal of a complex connection. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Miao, B.; Wang, H.; Wang, C.; Zhang, S.W. Splenic abscess caused by Streptococcus gallolyticus subsp. pasteurianus as presentation of a pancreatic cancer. J. Clin. Microbiol. 2013, 51, 4249–4251. [Google Scholar] [PubMed]
- Corredoira, J.; Alonso, M.P.; García-Garrote, F.; Garcia-Pais, M.J.; Coira, A.; Rabunal, R.; Gonzalez-Ramirez, A.; Pita, J.; Matesanz, M.; Velasco, D.; et al. Streptococcus bovis group and biliary tract infections: An analysis of 51 cases. Clin. Microbiol. Infect. 2013, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Matesanz, M.; Rubal, D.; Iñiguez, I.; Rabuñal, R.; García-Garrote, F.; Coira, A.; García-País, M.J.; Pita, J.; Rodriguez-Macias, A. Is Streptococcus bovis a urinary pathogen? Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, L.L.; Bisqaard, M.; Son, N.T.; Trung, N.V.; An, H.M.; Dalsgaard, A. Enterococcus and Streptococcus spp. associated with chronic and self-medicated urinary tract infections in Vietnam. BMC Infect. Dis. 2012, 12, 320. [Google Scholar] [CrossRef] [PubMed]
- Sheng, W.H.; Chuang, Y.; Teng, L.J.; Hsueh, P.P. Bacteraemia due to Streptococcus gallolyticus subspecies pasteurianus is associated with digestive tract malignancies and resistance to macrolides and clindamycin. J. Infect. Chemother. 2014, 69, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Chan, H.M.; Ho, P.L.; Wong, H.K.; Leung, J.N.; Tsoi, W.C.; Lin, C.K. Long-term clinical outcomes after Streptococcus bovis isolation in asymptomatic blood donors in Hong Kong. Transfusion 2013, 53, 2207–2210. [Google Scholar] [PubMed]
- Saegeman, V.; Cossey, V.; Loens, K.; Schuermans, A.; Glaser, P. Streptococcus gallolyticus subsp. pasteurianus infection in a neonatal intensive care unit. Pediatr. Infect. Dis. J. 2016. [Google Scholar] [CrossRef]
- Samkar, A.; Brouwer, M.C.; Pannekoek, Y.; Ende, A.; Beek, D. Streptococcus gallolyticus meningitis in adults: Report of five cases and review of the literature. Clin. Microbiol. Infect. 2015, 21, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- García-Pais, M.J.; Rabuñal, R.; Corredoira, J. Alonso MPSpondylodiscitis due to Streptococcus gallolyticus subsp. pasteurianus. Enferm. Infecc. Microbiol. Clin. 2016, 34, 141–142. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Takegawa, H.; Sakizono, K.; Nomoto, R.; Yamamoto, G.; Osawa, R. Transient bacteremia due to Streptococcus gallolyticus subsp. pasteurianus in a 3-year-old infant. Jpn. J. Infect. Dis. 2015, 68, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Eun, S.H.; Kim, E.C.; Seong, M.W.; Kim, Y.K. Neonatal invasive Streptococcus gallolyticus subsp. pasteurianus infection with delayed central nervous system complications. Korean J. Pediatr. 2015, 58, 33–36. [Google Scholar] [PubMed]
- Kimpe, A.; Decostere, A.; Martel, A.; Haesebrouck, F.; Devriese, A. Prevalence of antimicrobial resistance among pigeon isolates of Streptococcus gallolyticus, Escherichia coli and Salmonella enterica serotype Typhimurium. Avian Pathol. 2002, 31, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, R.; Huet, C.; Picherot, M.; Trieu-Cuot, P.; Trieu-Cuot, P.; Poyart, C. Genetic basis of antibiotic resistance in clinical isolates of Streptococcus gallolyticus (Streptococcus bovis). Antimicrob. Agents Chemother. 2005, 49, 1646–1648. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.C.; Hsueh, P.R.; Chen, H.J.; Tseng, S.P.; Cheng, P.Y.; Teng, L.J. The erm(T) gene is flanked by IS1216V in inducible erythromycin-resistant Streptococcus gallolyticus subsp. pasteurianus. Antimicrob. Agents Chemother. 2005, 49, 4347–4350. [Google Scholar] [CrossRef] [PubMed]
- Teng, L.J.; Hsueh, P.R.; Ho, S.W.; Luh, K.T. High prevalence of inducible erythromycin resistance among Streptococcus bovis isolates in Taiwan. Antimicrob. Agents Chemother. 2001, 45, 3362–3365. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.; Hidalgo, M.; Díaz, L.; Rincon, S.; Moreno, J.; Vanegas, N.; Castaneda, E. Characterization of macrolide resistance in Gram-positive cocci from Colombian hospitals: A countrywide surveillance. Int. J. Infect. Dis. 2007, 11, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Wierzbowski, A.K.; Boyd, D.; Mulvey, M.; Hoban, D.J.; Zhanel, G.G. Expression of the mef(E) gene encoding the macrolide efflux pump protein increases in Streptococcus pneumoniae with increasing resistance to macrolides. Antimicrob. Agents Chemother. 2005, 49, 4635–4640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tatsunol, I.; Okada, R.; Hata, N.; Matsumoto, M.; Isaka, M.; Isobe, K.; Haseqawa, T. Predominant role of msr(D) over mef(A) in macrolide-resistance in Streptococcus pyogenes. Microbiology 2015, 162, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.A.; Chochua, S.; Jackson, D.; Beall, B.; McGee, L. Mobile elements and chromosomal changes associated with MLS resistance phenotypes of invasive pneumococci recovered in the United States. Microb. Drug Resist. 2015, 21, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Dunkle, J.A.; Xiong, L.; Mankin, A.S.; Cate, J.H. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl. Acad. Sci. USA 2010, 107, 17152–17157. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 2002, 34, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Rosato, A.; Vicarini, H.; Leclercq, R. Inducible or constitutive expression of resistance in clinical isolates of streptococci and enterococci cross-resistant to erythromycin and lincomycin. J. Antimicrob. Chemother. 1999, 43, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twentieth Informational Supplement M100-S20; CLSI: Wayne, PA, USA, 2010. [Google Scholar]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard-Ninth Edition, M07-A9; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Moore, S.D.; Sauer, R.T. Revisiting the mechanism of macrolide-antibiotic resistance mediated by ribosomal protein L22. Proc. Natl. Acad. Sci. USA 2008, 105, 18261–18265. [Google Scholar] [CrossRef] [PubMed]
- Zaman, S.; Fitzpatrick, M.; Lindahl, L.; Zengel, J. Novel mutations in ribosomal proteins L4 and L22 that confer erythromycin resistance in Escherichia coli. Mol. Microbiol. 2007, 66, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.A.; Woo, P.C.Y.; To, A.P.C.; Lau, S.K.P.; Wong, S.S.Y.; Yuen, K.Y. Geographical difference of disease association in Streptococcus bovis bacteraemia. J. Med. Microbiol. 2003, 52, 903–908. [Google Scholar] [CrossRef] [PubMed]
- DiPersio, L.P.; DiPersio, J.R. Identification of an erm(T) gene in strains of inducibly clindamycin-resistant group B Streptococcus. Diagn. Microbiol. Infect. Dis. 2007, 57, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Skinner, R.; Cundliffe, E.; Schmidt, F.J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J. Biol. Chem. 1983, 258, 12702–12706. [Google Scholar] [PubMed]
- Weisblum, B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 1995, 39, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S.; Weisblum, B. Posttranscriptional modification of mRNA conformation: Mechanism that regulates erythromycin-induced resistance. Proc. Natl. Acad. Sci. USA 1980, 77, 7079–7083. [Google Scholar] [CrossRef] [PubMed]
- Arenz, S.; Ramu, H.; Gupta, P.; Berninghausen, O.; Beckmann, R.; Vazquez-Laslop, N.; Mankin, A.S.; Wilson, D.N. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat. Commun. 2014, 5, 3501. [Google Scholar] [CrossRef] [PubMed]
- Lovett, P.S.; Rogers, E.J. Ribosome regulation by the nascent peptide. Microbiol. Rev. 1996, 60, 366–385. [Google Scholar] [PubMed]
- Wilson, K.H.; Blitchinqton, R.; Greene, R.C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 1990, 28, 1942–1946. [Google Scholar] [PubMed]
- Prunier, A.L.; Malbruny, B.; Tandé, D.; Picard, B.; Leclercq, R. Clinical isolates of Staphylococcus aureus with ribosomal mutations conferring resistance to macrolides. Antimicrob. Agents Chemother. 2002, 46, 3054–3056. [Google Scholar] [CrossRef] [PubMed]
| Antibiotics | MIC (mg/L) of Subspecies | |||||||
|---|---|---|---|---|---|---|---|---|
| AL 101002 | GX 130304 | GX 130307 | GX 130630 | GX 130723 | GX 130809 | ATCC 43144 | ATCC 29213 | |
| Erythromycin | 1024 | 1280 | 1280 | 1280 | 1280 | 1024 | <0.25 | 0.25 |
| Clarithromycin | 512 | 512 | 512 | 512 | 512 | 512 | <0.25 | 0.25 |
| Lincomycin | 128 | >256 | >256 | 256 | 128 | 256 | 0.5 | 0.5 |
| Tetracyclin | 25 | 128 | 128 | 25 | 256 | 256 | 1 | 0.5 |
| Chloramphenicol | 2 | 32 | 32 | 2 | 2 | 2 | 2 | 2 |
| Levofloxacin | 8 | 8 | 4 | 8 | 4 | 8 | 4 | 0.25 |
| Gentamycin | 1 | 8 | 8 | 1 | 512 | 0.5 | 8 | 0.5 |
| Penicillin | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 2 |
| Cefotaxime | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.125 | <0.0625 | 2 |
| Vancomycin | <1 | <1 | <1 | <1 | <1 | <1 | <1 | 2 |
| Mutation in 23S rRNA | Found in Isolates | Location in 23S rRNA | MIC (mg/L) | ||
|---|---|---|---|---|---|
| S. pasteurianus Numbering | E. coli Numbering | Erythromycin | Clarithromycin | ||
| Wild Type | ATCC 43144 | 1 | 1 | ||
| U2824C | U2828C | AL101002 | Domain VI | 1 | 4 |
| GX130307 | |||||
| GX130630 | |||||
| GX130809 | |||||
| C2876U | C2880U | GX130723 | Domain VI | 2 | 8 |
| U2824C + C2876U | U2828C + C2880U | GX130723 | Domain VI | 4 | 16 |
| G1380U + A1409G + A1515G | G1355U + A1384G + A1490G | GX130304 | Domain III | 32 | 32 |
| erm Genes and Plasmids | MICs (mg/L) | ||
|---|---|---|---|
| Erythromycin | Clarithromycin | Lincomycin | |
| pET28a | 1 | 1 | 128 |
| PHT01 | 1 | 1 | 128 |
| ermT in pET28a | 1024 | 512 | 512 |
| ermTL in pET28a | 512 | 256 | 512 |
| ermB in pET28a | 1024 | 128 | 512 |
| ermBL in pET28a | 512 | 8 | 512 |
| ermT in PHT01 | 128 | 16 | – |
| ermTL in PHT01 | 16 | 4 | – |
| ermB in pET28a + ermT in PHT01 | 2048 | >1024 | 2560 |
| ermBL in pET28a + ermTL in PHT01 | 1024 | 1024 | 2048 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Cai, C.; Chen, J.; Cheng, C.; Cheng, G.; Hu, X.; Liu, C. Inducible Expression of both ermB and ermT Conferred High Macrolide Resistance in Streptococcus gallolyticus subsp. pasteurianus Isolates in China. Int. J. Mol. Sci. 2016, 17, 1599. https://doi.org/10.3390/ijms17101599
Li M, Cai C, Chen J, Cheng C, Cheng G, Hu X, Liu C. Inducible Expression of both ermB and ermT Conferred High Macrolide Resistance in Streptococcus gallolyticus subsp. pasteurianus Isolates in China. International Journal of Molecular Sciences. 2016; 17(10):1599. https://doi.org/10.3390/ijms17101599
Chicago/Turabian StyleLi, Meixia, Chao Cai, Juan Chen, Changwei Cheng, Guofu Cheng, Xueying Hu, and Cuiping Liu. 2016. "Inducible Expression of both ermB and ermT Conferred High Macrolide Resistance in Streptococcus gallolyticus subsp. pasteurianus Isolates in China" International Journal of Molecular Sciences 17, no. 10: 1599. https://doi.org/10.3390/ijms17101599