Diatom Valve Three-Dimensional Representation: A New Imaging Method Based on Combined Microscopies
Abstract
:1. Introduction
2. Results and Discussion
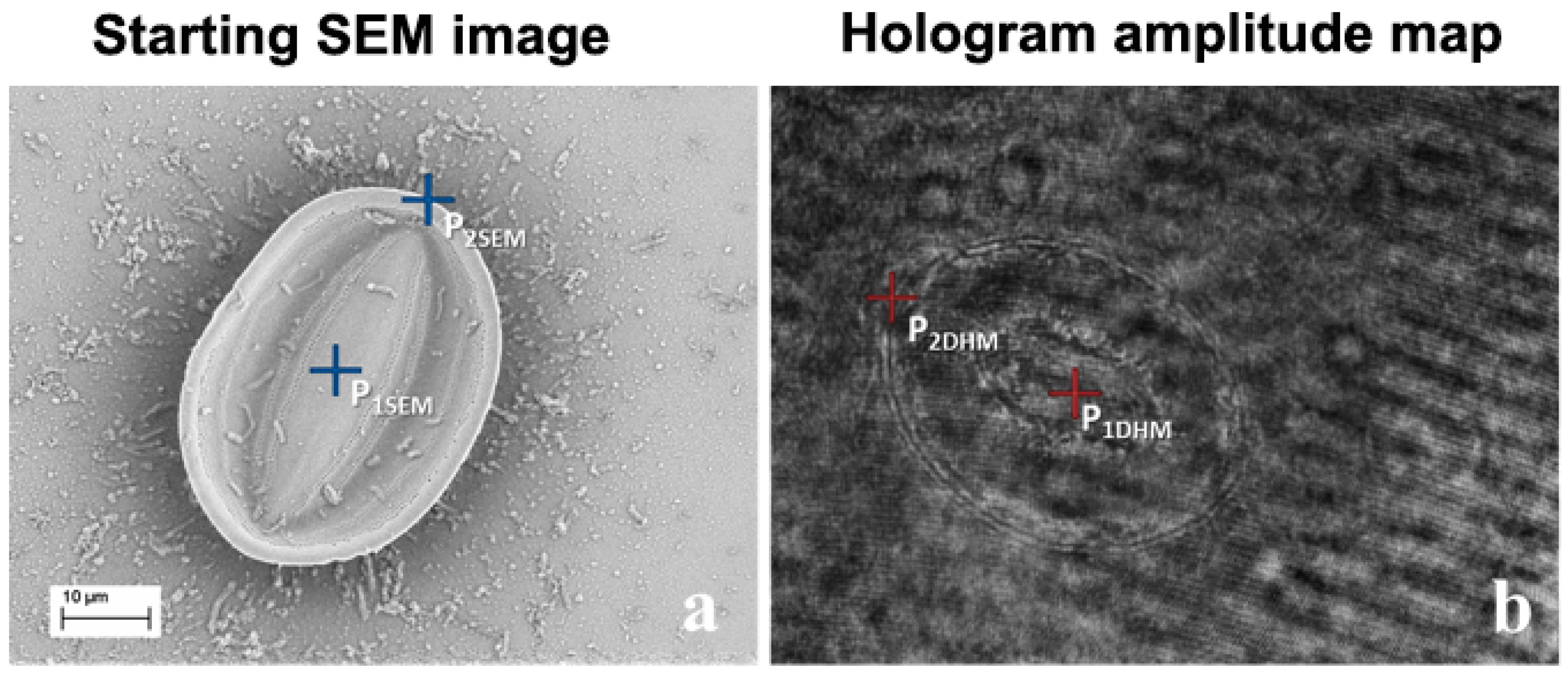
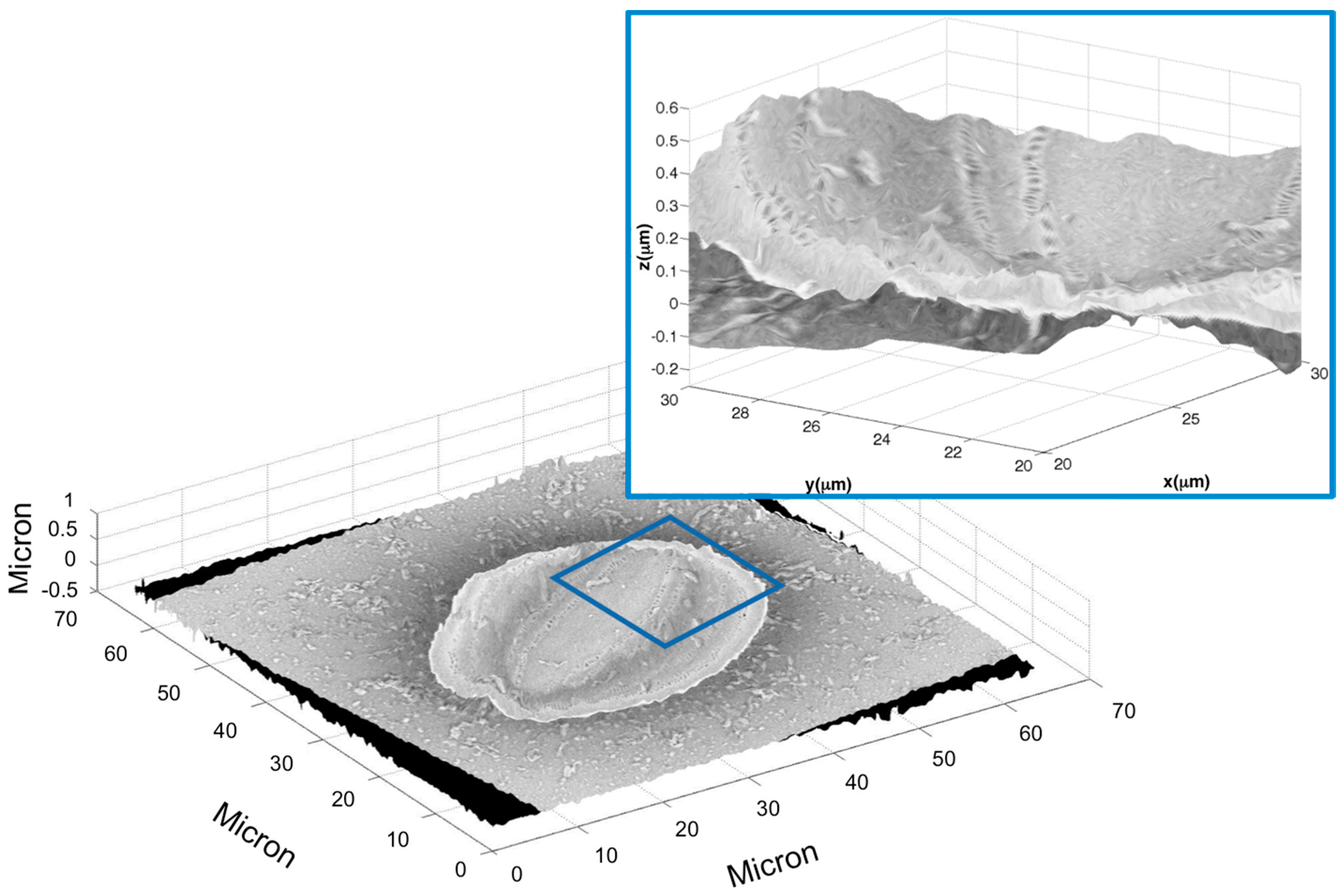
2.1. Example 1: Creating a 3D Model of an Arachnoidiscus Diatom Valve
2.2. Example 2: Creating a 3D Model of a Cocconeis Diatom Valve
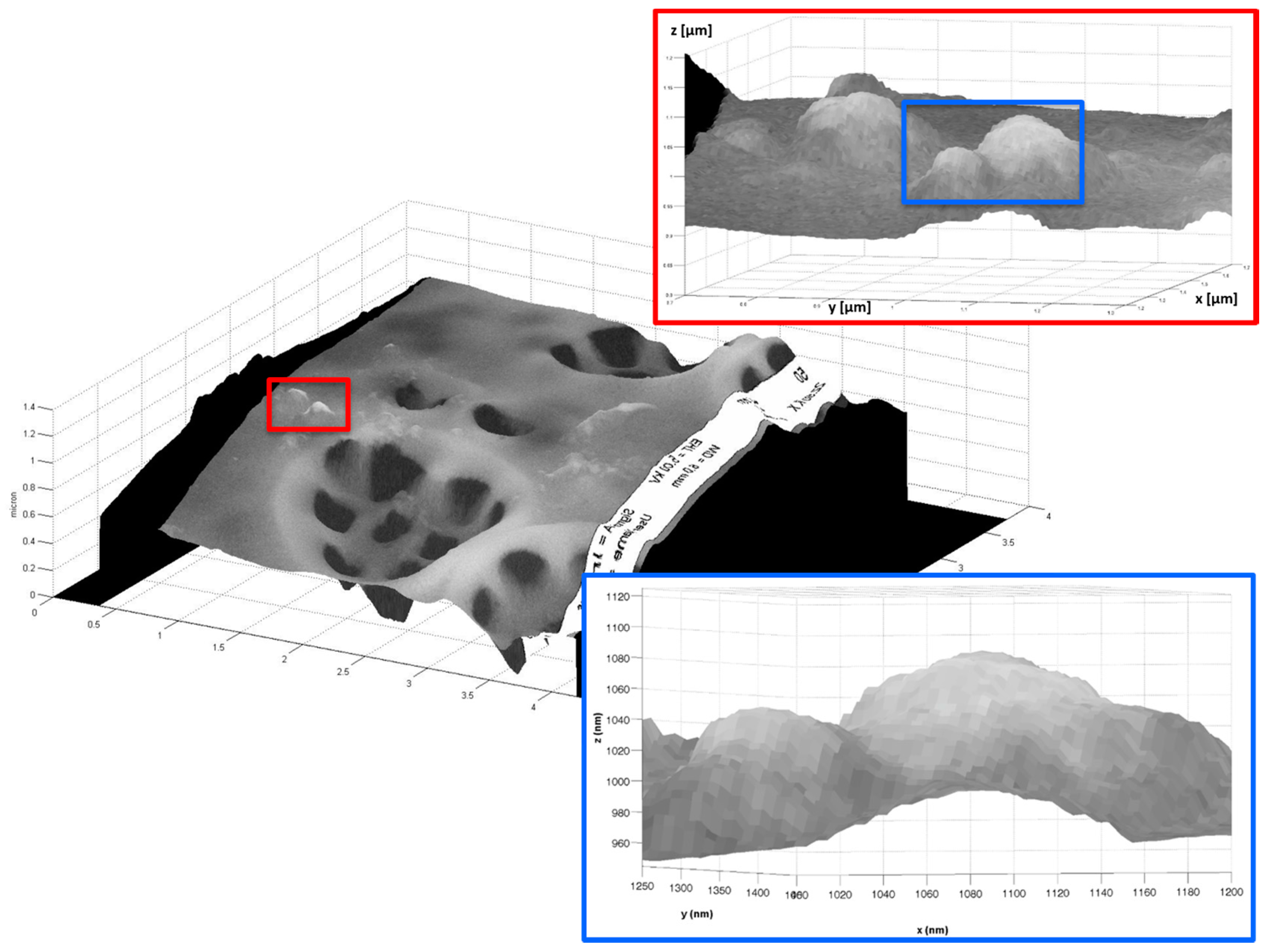
2.3. Example 3: Creating a 3D Image of a Porous of Coscinodiscus wailesii Diatom
3. Materials and Methods
3.1. Sample Origin and Preparation
3.2. Digital Holography Microscopy
3.3. Scanning Electron Microscopy
3.4. Atomic Force Microscopy
3.5. Mathematical Procedure
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- De Stefano, M.; de Stefano, L. Nanostructures in diatom frustules: Functional morphology of valvocopulae in Cocconeidacean monoraphid taxa. J. Nanosci. Nanotechnol. 2005, 5, 15–24. [Google Scholar] [CrossRef] [PubMed]
- De Tommasi, E.; Rea, I.; de Stefano, L.; Dardano, P.; di Caprio, G.; Ferrara, M.A.; Coppola, G. Optics with diatoms: Towards efficient, bioinspired photonic devices at the micro-scale. In Proceedings of the SPIE Optical Methods for Inspection, Characterization, and Imaging of Biomaterials, Munich, Germany, 13–16 May 2013.
- Ferrara, M.A.; Dardano, P.; de Stefano, L.; Rea, I.; Coppola, G.; Rendina, I.; Congestri, R.; Antonucci, A.; de Stefano, M.; de Tommasi, E. Optical properties of diatom nanostructured biosilica in Arachnoidiscus sp: Micro-optics from mother nature. PLoS ONE 2014, 9, e103750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Ostadi, H.; Jiang, K. Three-dimensional surface reconstruction of diatomaceous frustules. Anal. Biochem. 2010, 403, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Nahrendorf, M.; Badea, C.; Hedlund, L.W.; Figueiredo, J.-L.; Sosnovik, D.E.; Johnson, G.A.; Weissleder, R. High-resolution imaging of murine myocardial infarction with delayed-enhancement cine micro-CT. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H3172–H3178. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, R.; Nicastro, D.; Mastronarde, D. New views of cells in 3D: An introduction to electron tomography. Trends Cell Biol. 2005. [Google Scholar] [CrossRef] [PubMed]
- Pierson, J.; Vos, M.; McIntosh, J.R.; Peters, P.J. Perspectives on electron cryo-tomography of vitreous cryo-sections. J. Electron. Microsc. 2011, 60, S93–S100. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Liu, Z.; Georgieva, D.; Kuil, M.E.; Abrahams, J.P. A novel approximation method of CTF amplitude correction for 3D single particle reconstruction. Ultramicroscopy 2010, 110, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Marinello, F.; Bariani, P.; Savio, E.; Horsewell, A.; de Chiffre, L. Critical factors in SEM 3D stereo microscopy. Meas. Sci. Technol. 2008. [Google Scholar] [CrossRef]
- Hildebrand, M.; Kim, S.; Shi, D.; Scott, K.; Subramaniam, S. 3D imaging of diatoms with Ion-abrasion scanning electron microscopy. J. Struct. Biol. 2009, 166, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Friedrichs, L.; Maier, M.; Hamm, C. A new method for exact three-dimensional reconstructions of diatom frustules. J. Microsc. 2012, 248, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Dardano, P.; Ferrara, M.A.; de Tommasi, E.; Coppola, G.; Rea, I.; Rendina, I.; de Stefano, L.; di Caprio, G. Method for the Reconstruction of a Three-Dimensional Model of a Microscopic Object. Italian Patent MI2014A000126, 30 January 2014. [Google Scholar]
- Simonsen, R.; Hustedt, F. Atlas and Catalogue of the Diatom Types of Friedrich Hustedt; J. Cramer: Berlin, Germany, 1987; pp. 20–32. [Google Scholar]
- Round, F.E.; Crawford, R.M.; Mann, D.G. The Diatoms: Biology and Morphology of the Genera; Cambridge University Press: Cambridge, UK, 1990; pp. 60–93. [Google Scholar]
- Maibohm, C.; Friis, S.M.M.; Ellegaard, M.; Rottwitt, K. Interference patterns and extinction ratio of the diatom Coscinodiscus granii. Opt. Express 2015, 23, 9543–9548. [Google Scholar] [CrossRef] [PubMed]
- De Tommasi, E.; de Luca, A.C.; Lavanga, L.; Dardano, P.; de Stefano, M.; de Stefano, L.; Langella, C.; Rendina, I.; Dholakia, K.; Mazilu, M. Biologically enabled sub-diffractive focusing. Opt. Express 2014, 22, 27214–27227. [Google Scholar] [CrossRef] [PubMed]
- Von Quillfeldt, C.H. Common diatom species in Arctic spring blooms: Their distribution and abundance. Bot. Mar. 2000, 43, 499–516. [Google Scholar] [CrossRef]
- Di Caprio, G.; Coppola, G.; de Stefano, L.; de Stefano, M.; Antonucci, A.; Congestri, R.; Tommasi, E.D. Shedding light on diatom photonics by means of digital holography. J. Biophotonics 2014, 7, 341–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnars, U.; Jüptner, W.P.O. Digital recording and numerical reconstruction of holograms. Meas. Sci. Technol. 2002, 13, R85–R101. [Google Scholar] [CrossRef]




© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrara, M.A.; De Tommasi, E.; Coppola, G.; De Stefano, L.; Rea, I.; Dardano, P. Diatom Valve Three-Dimensional Representation: A New Imaging Method Based on Combined Microscopies. Int. J. Mol. Sci. 2016, 17, 1645. https://doi.org/10.3390/ijms17101645
Ferrara MA, De Tommasi E, Coppola G, De Stefano L, Rea I, Dardano P. Diatom Valve Three-Dimensional Representation: A New Imaging Method Based on Combined Microscopies. International Journal of Molecular Sciences. 2016; 17(10):1645. https://doi.org/10.3390/ijms17101645
Chicago/Turabian StyleFerrara, Maria Antonietta, Edoardo De Tommasi, Giuseppe Coppola, Luca De Stefano, Ilaria Rea, and Principia Dardano. 2016. "Diatom Valve Three-Dimensional Representation: A New Imaging Method Based on Combined Microscopies" International Journal of Molecular Sciences 17, no. 10: 1645. https://doi.org/10.3390/ijms17101645











