Identification, Molecular Cloning of IL-1β and Its Expression Profile during Nocardia seriolae Infection in Largemouth Bass, Micropterus salmoides
Abstract
:1. Introduction
2. Results
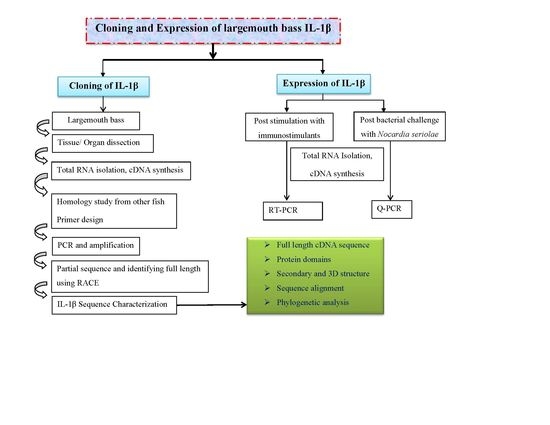
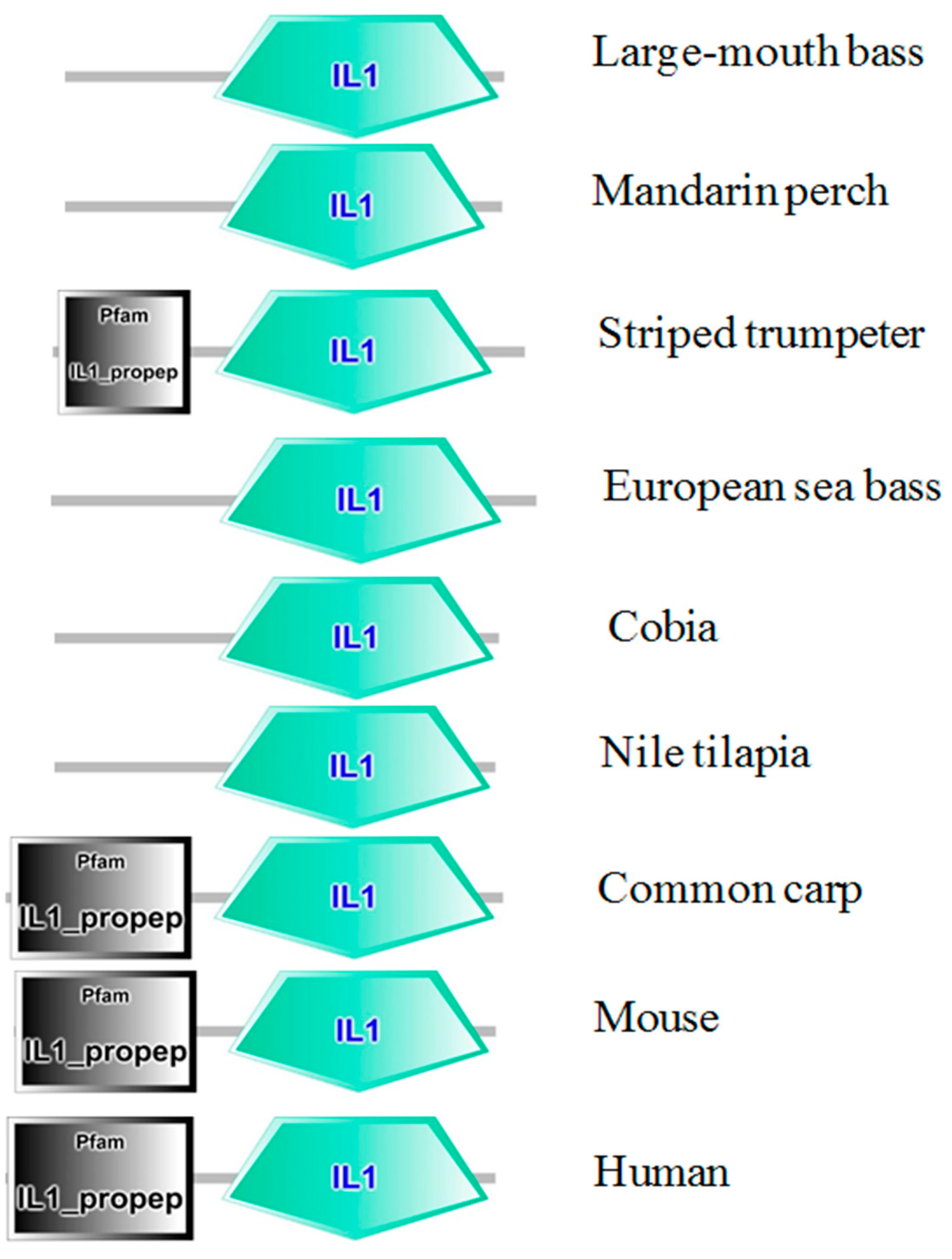
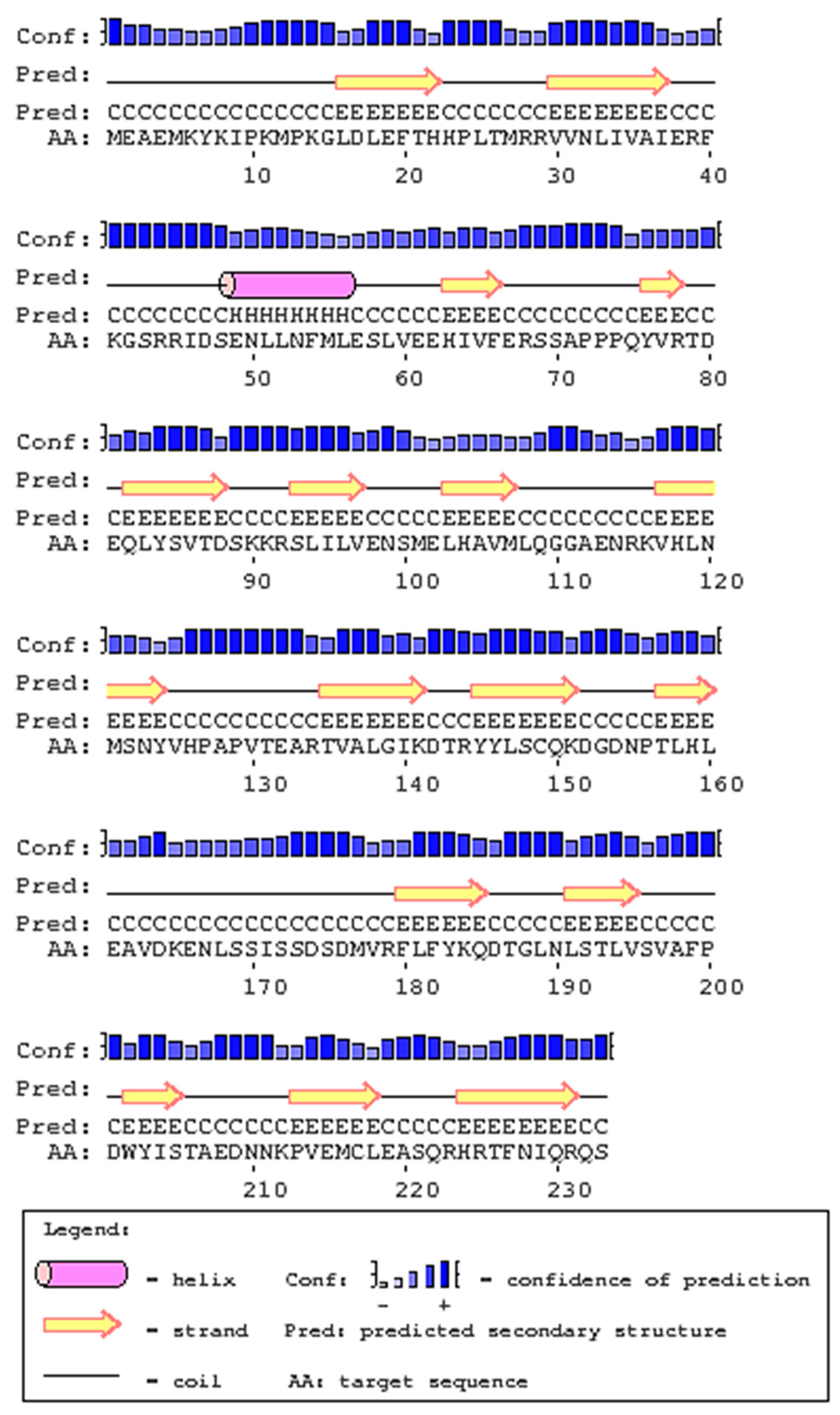
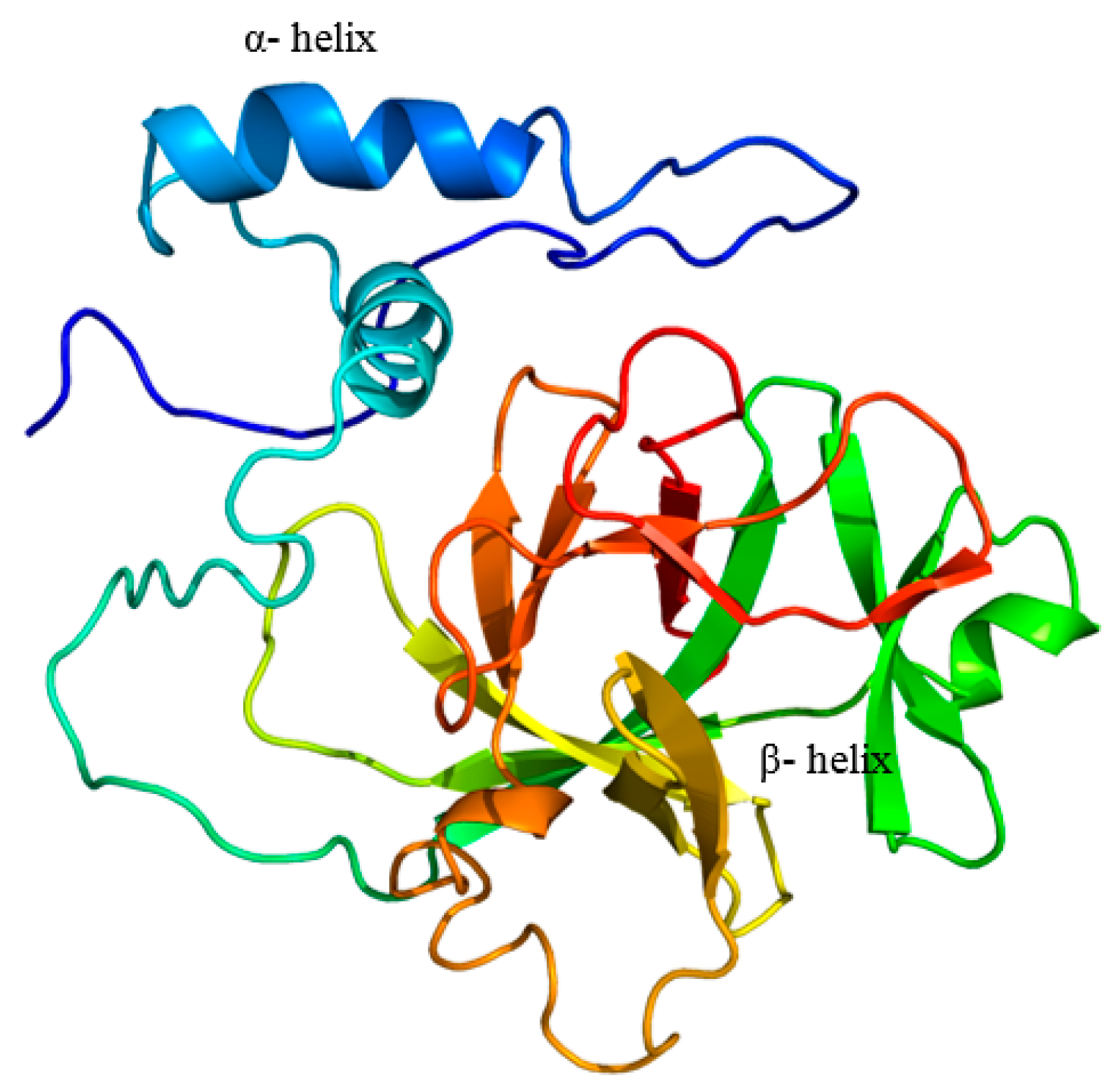
2.1. Cloning and Analysis of Full-Length IL-1β mRNA
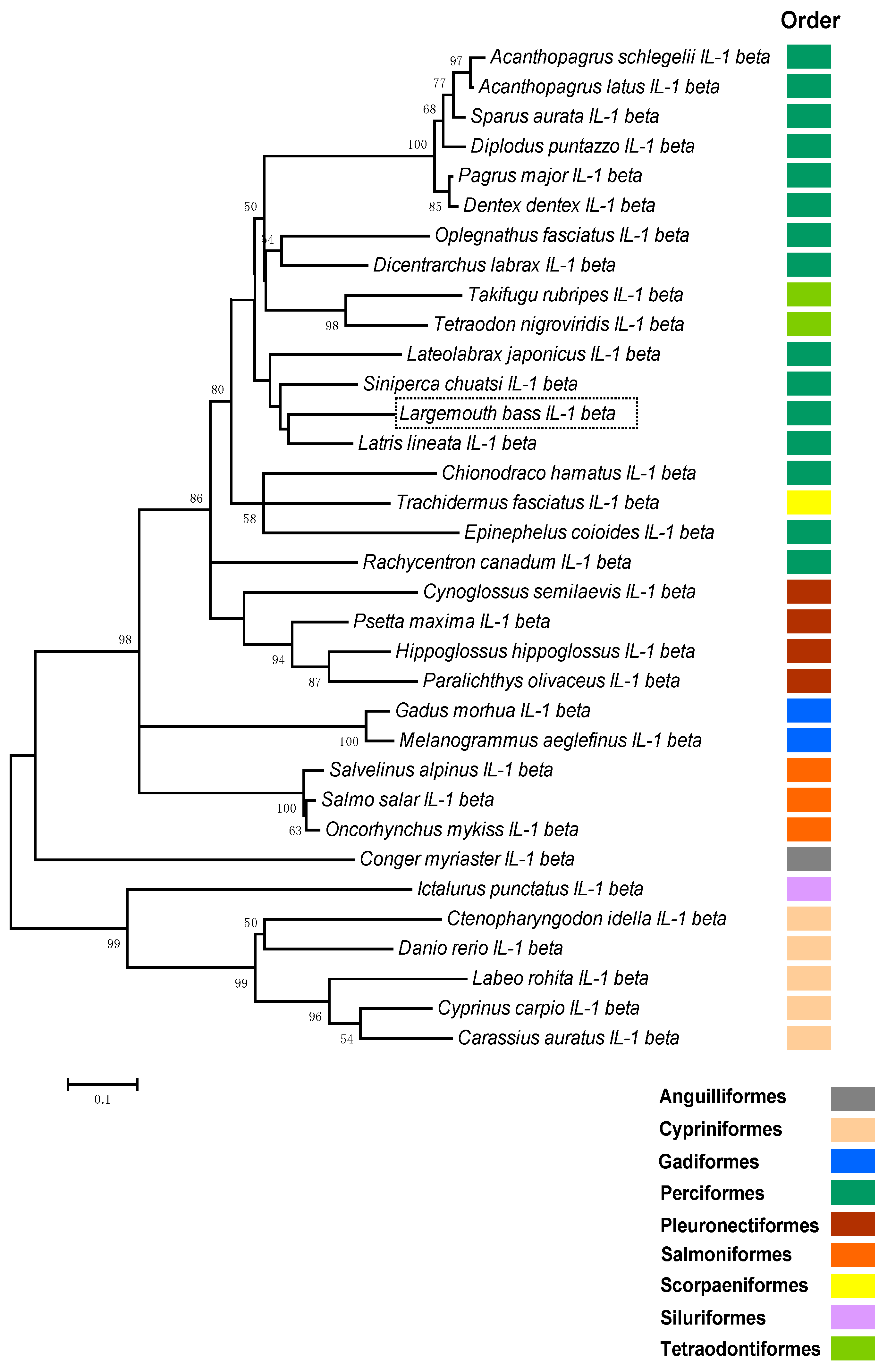
2.2. Phylogenetic Analysis of Teleosts IL-1β
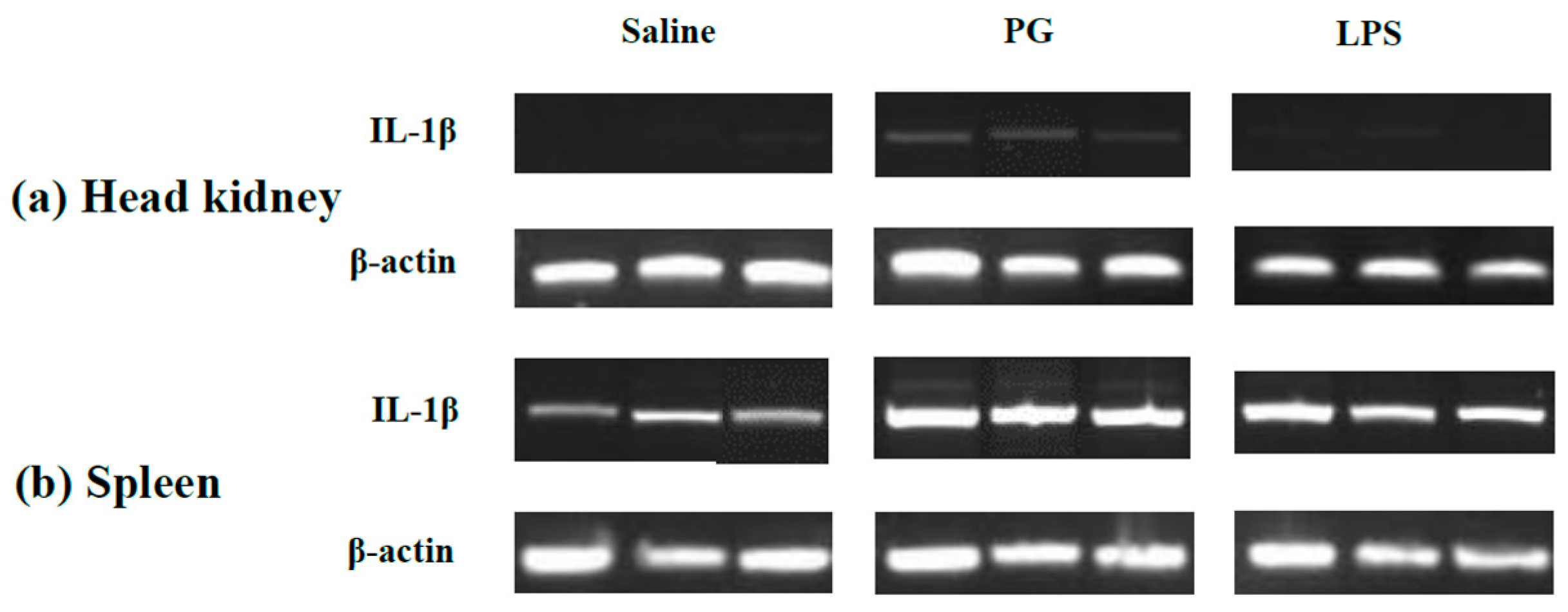
2.3. Expression Profile of LMBIL-1β in Spleen and Head Kidney after LPS and Peptidoglycan Stimulation
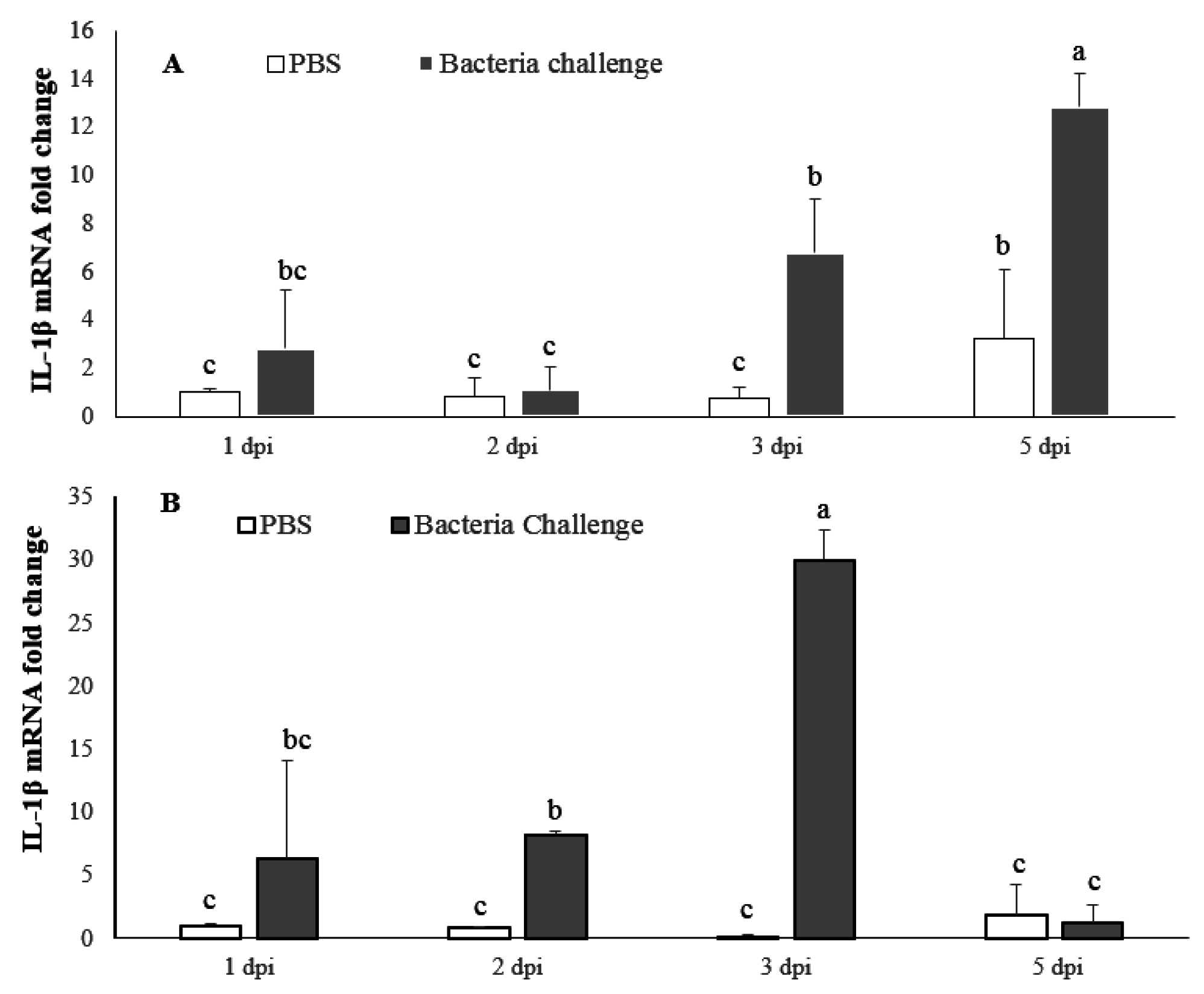
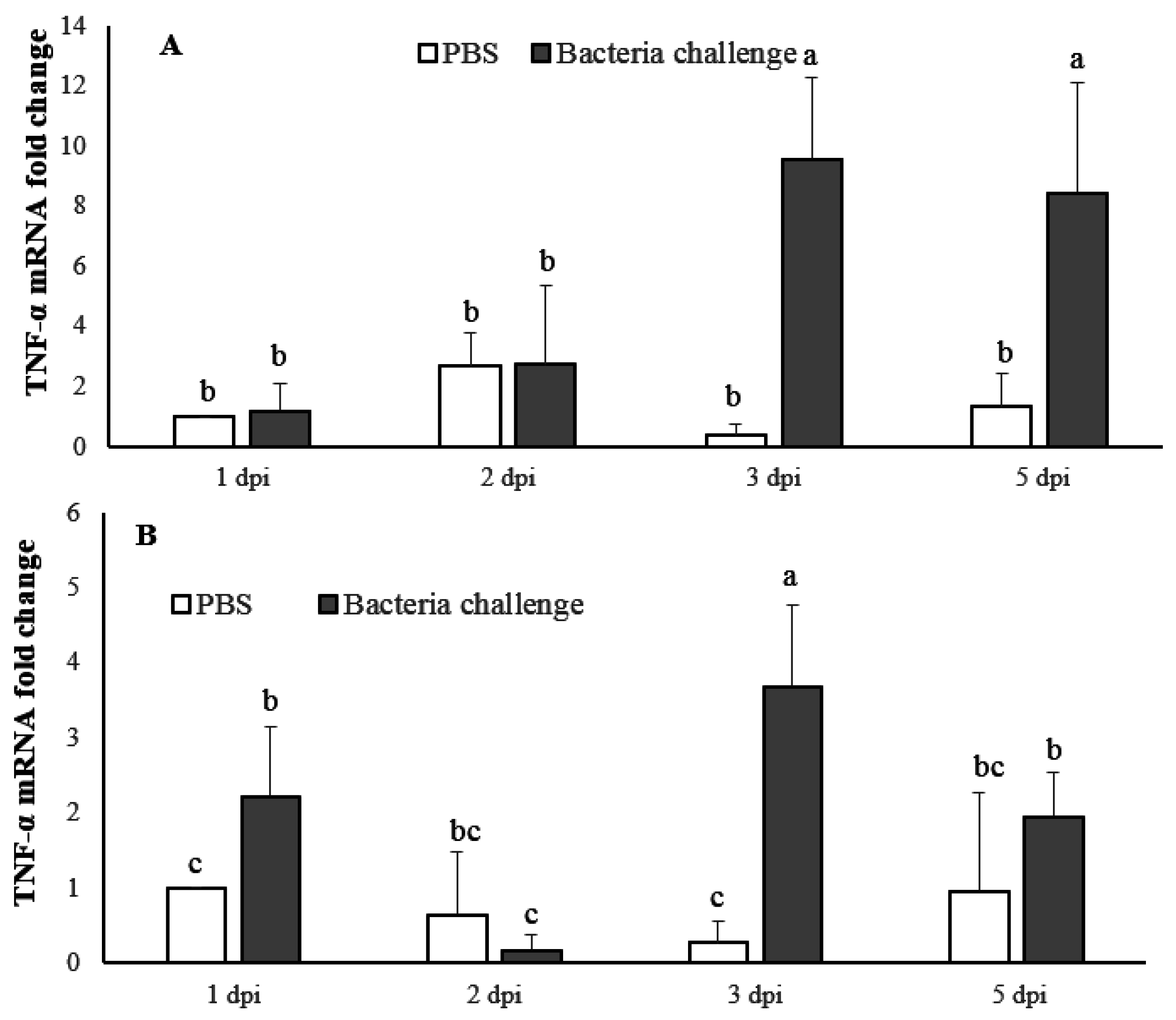
2.4. Expression Profile of LMBIL-1β and TNF-α in Spleen and Head Kidney after Nocardia Seriolae Challenge
2.4.1. LMBIL-1β mRNA Expression
2.4.2. TNF-α mRNA Expression
3. Discussion
4. Materials and Methods
4.1. Animal Maintenance
4.2. Primer Design and RT-PCR to Isolate Partial IL-1β Sequence from Largemouth Bass
4.3. Sequencing and Full-Length cDNA Cloning
4.4. In-Silico Analysis of Largemouth Bass IL-1β Sequence
4.5. Phylogenetic Analysis
4.6. Fish IL-1β Induction and RNA Extraction
4.7. IL-1β Expressional Profiling by RT-PCR
4.8. Isolation, Cultivation and Challenge with Nocardia Seriolae
4.9. Real Time Polymerase Chain Reaction
4.10. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dinarello, C.A. Biology of interleukin 1. FASEB J. 1988, 2, 108–115. [Google Scholar]
- Dinarello, C.A. IL-1: Discoveries, controversies and future directions. Eur. J. Immunol. 2010, 40, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Biologic basis for interleukin-1 in disease. Blood 1996, 87, 2095–2147. [Google Scholar] [PubMed]
- Howard, A.D.; Kostura, M.J.; Thornberry, N.; Ding, G.J.; Limjuco, G.; Weidner, J.; Salley, J.P.; Hogquist, K.A.; Chaplin, D.D.; Mumford, R.A. IL-1-converting enzyme requires aspartic acid residues for processing of the IL-1β precursor at two distinct sites and does not cleave 31-kDa IL-1 α. J. Immunol. 1991, 147, 2964–2969. [Google Scholar] [PubMed]
- Dinarello, C.A. The IL-1 family and inflammatory diseases. Clin. Exp. Rheumatol. 2002, 20, S1–S13. [Google Scholar] [PubMed]
- Lopez-Castejon, G.; Brough, D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011, 22, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Wasiliew, P.; Kracht, M. Interleukin-1 (IL-1) pathway. Sci. Signal. 2010, 3. [Google Scholar] [CrossRef] [PubMed]
- Clem, L.W.; Sizemore, R.C.; Ellsaesser, C.F.; Miller, N.W. Monocytes as accessory cells in fish immune responses. Dev. Comp. Immunol. 1985, 9, 803–809. [Google Scholar] [CrossRef]
- Sigel, M.M.; Hamby, B.A.; Huggins, E.M. Phylogenetic studies on lymphokines. Fish lymphocytes respond to human IL-1 and epithelial cells produce an IL-1 like factor. Vet. Immunol. Immunopathol. 1986, 12, 47–58. [Google Scholar] [CrossRef]
- Verburg-van Kemenade, B.M.; Weyts, F.A.; Debets, R.; Flik, G. Carp macrophages and neutrophilic granulocytes secrete an interleukin-1-like factor. Dev. Comp. Immunol. 1995, 19, 59–70. [Google Scholar] [CrossRef] [Green Version]
- Weyts, F.A.A.; Rombout, J.H.W.M.; Flik, G.; Verburg-Van Kemenade, B.M.L. A common carp (Cyprinus carpio) leucocyte cell line shares morphological and functional characteristics with macrophages. Fish Shellfish Immunol. 1997, 7, 123–133. [Google Scholar] [CrossRef]
- Secombes, C.J.; Bird, S.; Cunningham, C.; Zou, J. Interleukin-1 in fish. Fish Shellfish Immunol. 1999, 9, 335–343. [Google Scholar] [CrossRef]
- Zou, J.; Grabowski, P.S.; Cunningham, C.; Secombes, C.J. Molecular cloning of interleukin 1β from rainbow trout Oncorhynchus mykiss reveals no evidence of an ice cut site. Cytokine 1999, 11, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Watzke, J.; Schirmer, K.; Scholz, S. Bacterial lipopolysaccharides induce genes involved in the innate immune response in embryos of the zebrafish (Danio rerio). Fish Shellfish Immunol. 2007, 23, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Morrison, R.N.; Young, N.D.; Nowak, B.F. Description of an Atlantic salmon (Salmo salar L.) type II interleukin-1 receptor cDNA and analysis of interleukin-1 receptor expression in amoebic gill disease-affected fish. Fish Shellfish Immunol. 2012, 32, 1185–1190. [Google Scholar] [CrossRef] [PubMed]
- Vojtech, L.N.; Scharping, N.; Woodson, J.; Hansen, J.D. Roles of inflammatory caspases during processing of zebrafish interleukin-1β in Francisella noatunensis infection. Infect. Immun. 2012, 80, 2878–2885. [Google Scholar] [CrossRef] [PubMed]
- Morrison, R.N.; Zou, J.; Secombes, C.J.; Scapigliati, G.; Adams, M.B.; Nowak, B.F. Molecular cloning and expression analysis of tumor necrosis factor-α in amoebic gill disease (AGD)-affected Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 2007, 23, 1015–1031. [Google Scholar] [CrossRef] [PubMed]
- Bo, Y.X.; Song, X.H.; Wu, K.; Bo, H.; Sun, B.Y.; Liu, Z.J. Characterization of interleukin-1β as a proinflammatory cytokine in grass carp. Fish Shellfish Immunol. 2015, 46, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Kudo, T.; Hatai, K.; Seino, A. Nocardia seriolae sp. nov. Causing nocardiosis of cultured fish. Int. J. Syst. Evol. Microbiol. 1988, 38, 173–178. [Google Scholar] [CrossRef]
- Itano, T.; Kawakami, H.; Kono, T.; Sakai, M. Live vaccine trials against nocardiosis in yellowtail Seriola quinqueradiata. Aquaculture 2006, 261, 1175–1180. [Google Scholar] [CrossRef]
- Snieszko, S.F.; Bullock, G.L.; Dunbar, C.E.; Pettijohn, L.L. Nocardial infection in hatchery-reared fingerling rainbow trout (Salmo gairdneri). J. Bacteriol. 1964, 88, 1809–1810. [Google Scholar] [PubMed]
- Chen, S.C.; Tung, M.C.; Tsai, W.C. An epizootic in Formosa snake-head fish, Channa maculata Lacepede, caused by Nocardia asteroides in fresh water pond in southern Taiwan. COA Fish Ser. 1989, 15, 42–48. [Google Scholar]
- Chen, S.C.; Tung, M.C. An epizootic in largemouth bass, Micropterus salmoides, Lacepede caused by Nocardia asteroides in freshwater pond in southern Taiwan. J. Chin. Soc. Vet. Sci. 1991, 17, 15–22. [Google Scholar]
- Wang, G.L.; Xu, Y.J.; Jin, S.; Zhu, J.L.; Yuan, S.P. Nocardiosis in snakehead, Ophiocephalus argus cantor. Aquaculture. 2007, 271, 54–60. [Google Scholar] [CrossRef]
- Chen, S.C.; Lee, J.L.; Lai, C.C.; Gu, Y.W.; Wang, C.T.; Chang, H.U.; Tsai, K.H. Nocardiosis in sea bass, Lateolabrax japonicus. J. Fish Dis. 2000, 23, 299–307. [Google Scholar] [CrossRef]
- Wang, G.L.; Yuan, S.P.; Jin, S. Nocardiosis in large yellow croaker, Larimichthys crocea (Richardson). J. Fish Dis. 2005, 28, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.C.; Chen, S.D.; Tsai, M.A.; Weng, Y.J.; Chu, S.Y.; Chern, R.S.; Chen, S.C. Nocardia seriolae infection in the three striped tigerfish, Terapon jarbua (Forsskål). J. Fish Dis. 2009, 32, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Bird, S.; Zou, J.; Wang, T.; Munday, B.; Cunningham, C.; Secombes, C.J. Evolution of interleukin-1β. Cytokine Growth Factor Rev. 2002, 13, 483–502. [Google Scholar] [CrossRef]
- Lee, D.S.; Hong, S.H.; Lee, H.J.; Jun, L.J.; Chung, J.K.; Kim, K.H.; Jeong, H.D. Molecular cDNA cloning and analysis of the organization and expression of the IL-1β gene in the Nile tilapia, Oreochromis niloticus. Comp. Biochem. Physiol. A 2006, 143, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, K.; Shin, D.H.; Nakao, M.; Yano, T. Molecular cloning and expression analysis of carp (Cyprinus carpio) interleukin-1 β, high affinity immunoglobulin E Fc receptor γ subunit and serum amyloid A. Fish. Shellfish. Immunol. 2000, 10, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Mulder, I.E.; Wadsworth, S.; Secombes, C.J. Cytokine expression in the intestine of rainbow trout (Oncorhynchus mykiss) during infection with Aeromonas salmonicida. Fish Shellfish Immunol. 2007, 23, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Tanekhy, M.; Matsuda, S.; Itano, T.; Kawakami, H.; Kono, T.; Sakai, M. Expression of cytokine genes in head kidney and spleen cells of Japanese flounder (Paralichthys olivaceus) infected with Nocardia seriolae. Vet. Immunol. Immunopathol. 2009, 134, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Clark, M.S.; Secombes, C.J. Characterization, expression and promoter analysis of an interleukin 10 homologue in the puffer fish, Fugu rubripes. Immunogenetics 2003, 55, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Laing, K.J.; Wang, T.; Zou, J.; Holland, J.; Hong, S.; Bols, N.; Hirono, I.; Aoki, T.; Secombes, C.J. Cloning and expression analysis of rainbow trout Oncorhynchus mykiss tumor necrosis factor-α. Eur. J. Biochem. 2001, 268, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Savan, R.; Sakai, M. Genomics of fish cytokines. Comp. Biochem. Physiol. 2006, 1, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Kwang, J. Carp interleukin-1 β in the role of an immuno-adjuvant. Fish Shellfish Immunol. 2000, 10, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Buonocore, F.; Mazzini, M.; Forlenza, M.; Randelli, E.; Secombes, C.J.; Zou, J.; Scapigliati, G. Expression in Escherichia coli and purification of sea bass (Dicentrarchus labrax) interleukin 1β, a possible immunoadjuvant in aquaculture. Mar. Biotechnol. 2004, 6, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Secombes, C.J. Two peptides derived from trout IL-1β have different stimulatory effects on immune gene expression after intraperitoneal administration. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2009, 153, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Sternberg, M.J.E. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Itano, T.; Kawakami, H.; Kono, T.; Sakai, M. Detection of fish nocardiosis by loop-mediated isothermal amplification. J. Appl. Microbiol. 2005, 100, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2 (−∆∆Ct) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Species Name | Amino Acid Identity (%) |
|---|---|
| Mandarin perch | 73 |
| Striped trumpeter | 73 |
| Striped beak fish | 65 |
| European sea bass | 66 |
| Japanese sea bass | 65 |
| Nile tilapia | 63 |
| Gilthead seabream | 58 |
| Cobia | 61 |
| Orange spotted grouper | 55 |
| Fugu | 61 |
| Atlantic salmon | 53 |
| Rainbow trout | 54 |
| Common carp | 35 |
| Primer Name | Primer Sequence (5′–3′) | Application |
|---|---|---|
| FIL1BF | TGGAMYTKGAGATTDCMCA | Partial cloning |
| FIL1BR | AAAYCKYACCATGTCGCTG | |
| Universal Primer Mix (UPM) | Long 0.2 μM CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | RACE |
| Short 0.4 μM CTAATACGACTCACTATAGGGC | ||
| Nested Universal Primer Mix (NUP) | AAGCAGTGGTATCAACGCAGAGT | |
| LMBIL1B3R1 | GCATCAAAGACACACGTTACTACCTGTC | |
| LMBIL-1b_F | TGGACTTGGAGATTGCCCA | q-PCR |
| LMBIL-1b_R | AAACCGCACCATGTCGCTG |
| Primer Name | Primer Sequence (5′–3′) | Target Gene | Application |
|---|---|---|---|
| β-Actin375F | CCACCACAGCCGAGAGGGAA | β-actin | q-PCR |
| β-Actin375R | TCATGGTGGATGGGGCCAGG | ||
| IL-1βF | TTGCCATAGAGAGGTTTA | IL-1β | |
| IL-1βR | ACACTATATGCTCTTCCA | ||
| TNFα-F | CTAGTGAAGAACCAGATTGT | TNF-α | |
| TNFα-R | AGGAGACTCTGAACGATG |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ho, P.-Y.; Byadgi, O.; Wang, P.-C.; Tsai, M.-A.; Liaw, L.-L.; Chen, S.-C. Identification, Molecular Cloning of IL-1β and Its Expression Profile during Nocardia seriolae Infection in Largemouth Bass, Micropterus salmoides. Int. J. Mol. Sci. 2016, 17, 1670. https://doi.org/10.3390/ijms17101670
Ho P-Y, Byadgi O, Wang P-C, Tsai M-A, Liaw L-L, Chen S-C. Identification, Molecular Cloning of IL-1β and Its Expression Profile during Nocardia seriolae Infection in Largemouth Bass, Micropterus salmoides. International Journal of Molecular Sciences. 2016; 17(10):1670. https://doi.org/10.3390/ijms17101670
Chicago/Turabian StyleHo, Ping-Yueh, Omkar Byadgi, Pei-Chyi Wang, Ming-An Tsai, Li-Ling Liaw, and Shih-Chu Chen. 2016. "Identification, Molecular Cloning of IL-1β and Its Expression Profile during Nocardia seriolae Infection in Largemouth Bass, Micropterus salmoides" International Journal of Molecular Sciences 17, no. 10: 1670. https://doi.org/10.3390/ijms17101670