A Comprehensive MicroRNA Expression Profile of Liver and Lung Metastases of Colorectal Cancer with Their Corresponding Host Tissue and Its Prognostic Impact on Survival
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
2.2. miRNA Profiling
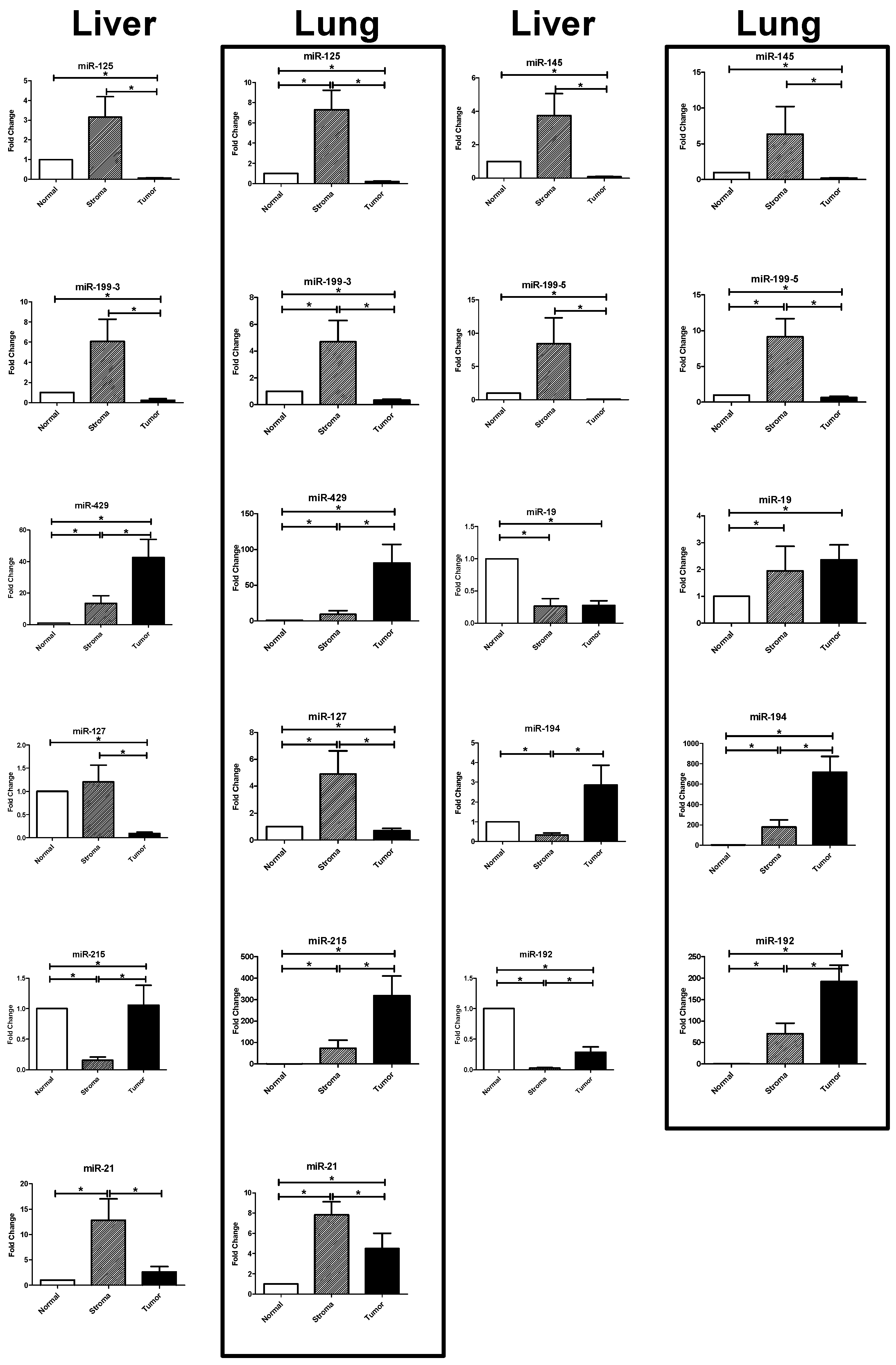
2.3. Results of the RT-PCR Analysis: Differential Expression in Tumor, Stroma, and Host Tissue
2.3.1. Upregulated miRNAs in the Tumoral Compartment of Liver and Lung Metastases
2.3.2. Upregulated miRNAs in the Stromal Compartment of Liver and Lung Metastases
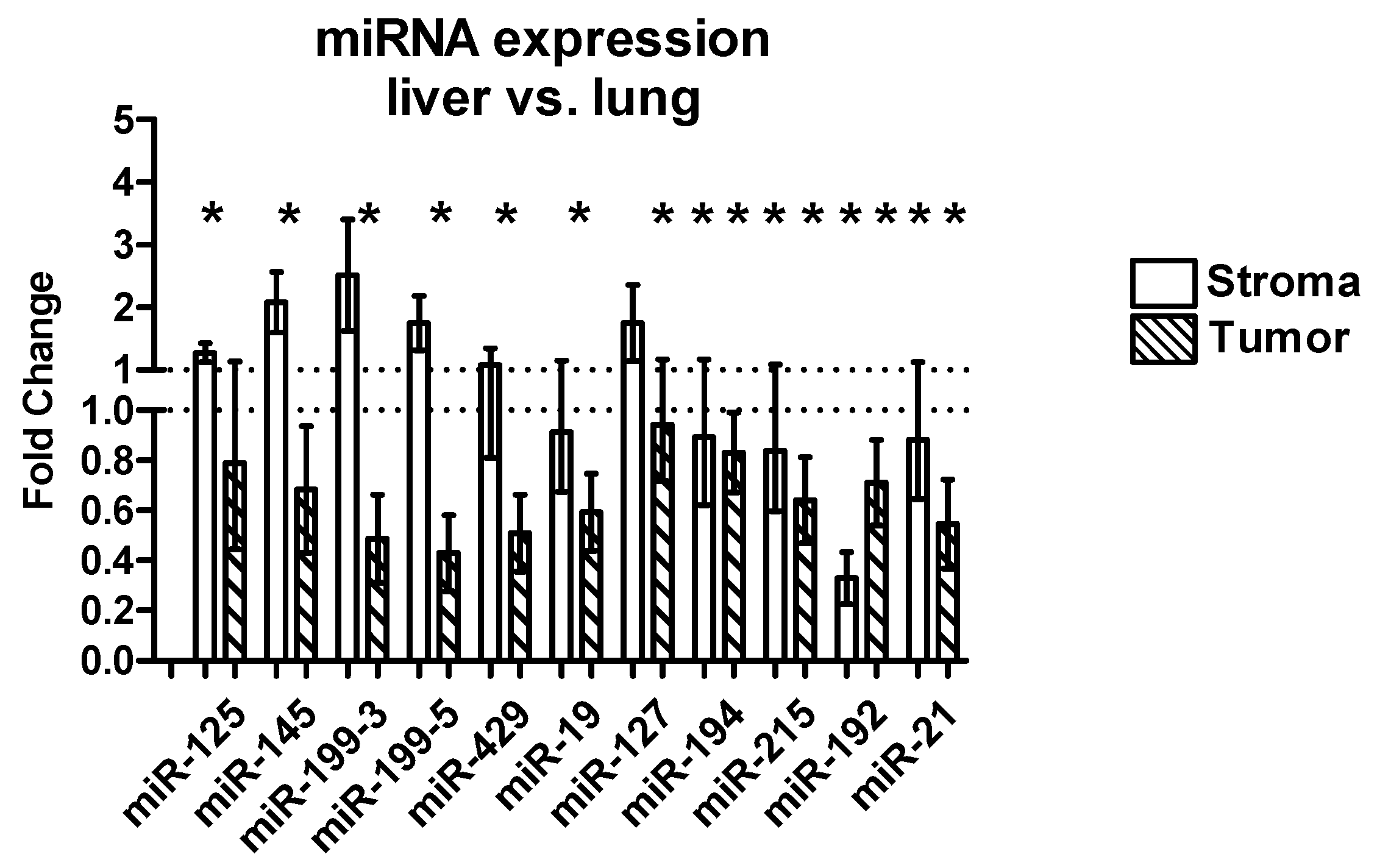
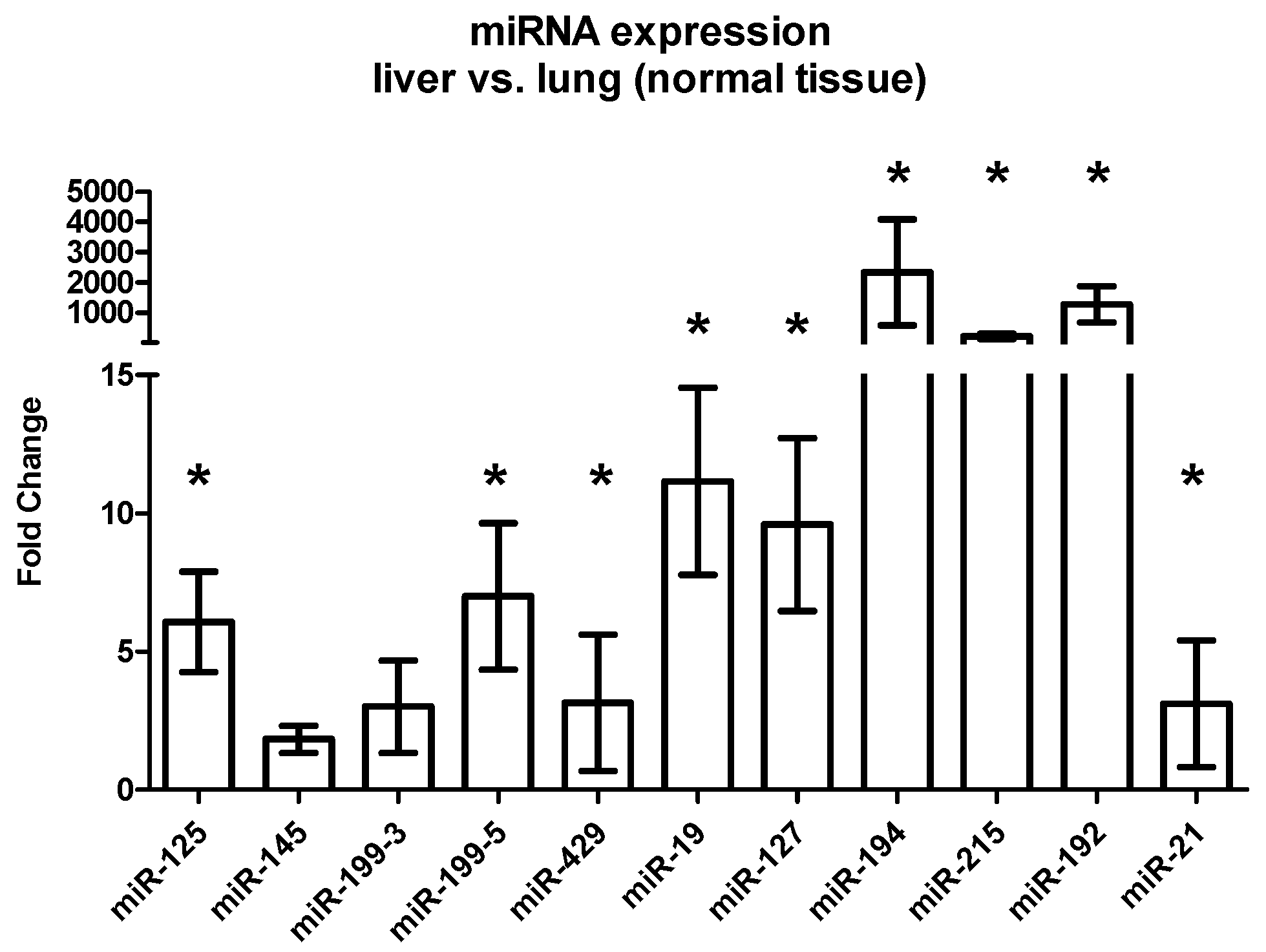
2.3.3. Differential Expression between Liver and Lung Metastases
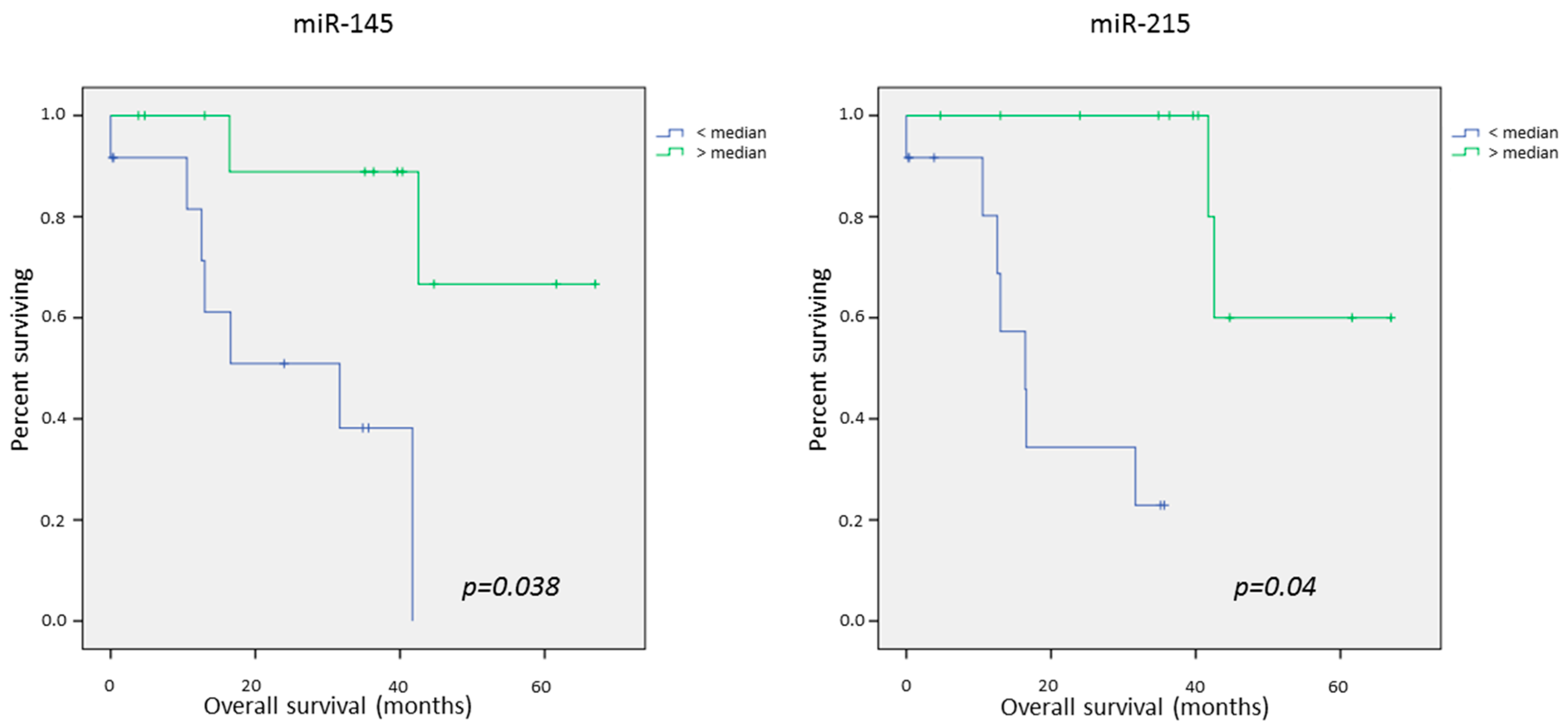
2.4. Survival Data
Univariate Survival Analysis
3. Discussion
4. Materials and Methods
4.1. Tumor Samples and Clinical Data
4.2. Clinical Specimens
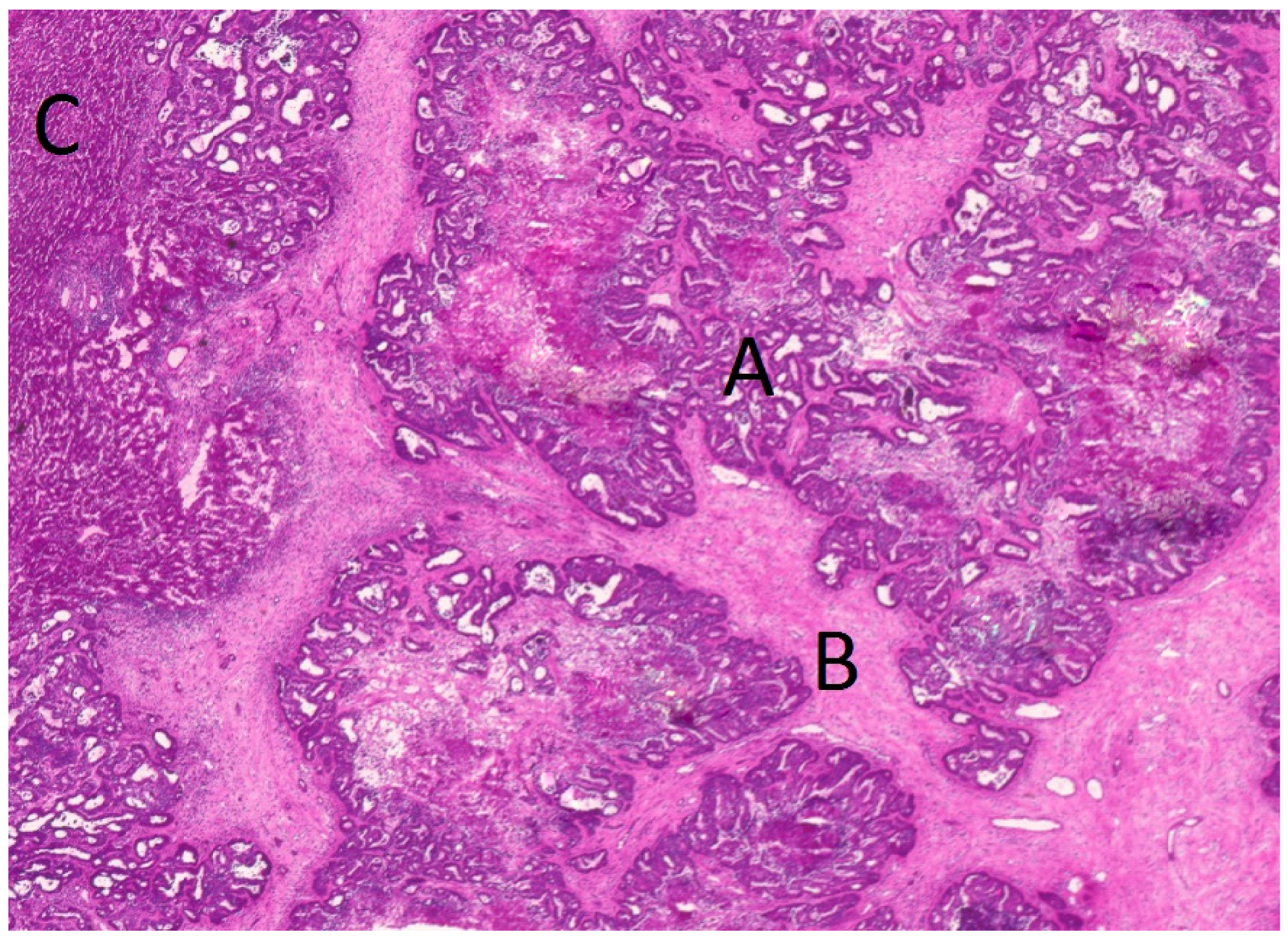
4.3. Tissue Preparation and Laser Microdissection
4.4. RNA Extraction
4.5. Microarray Analysis
4.6. Real-Time Quantitative PCR (qRT-PCR)
4.7. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Manfredi, S.; Lepage, C.; Hatem, C.; Coatmeur, O.; Faivre, J.; Bouvier, A.-M. Epidemiology and Management of Liver Metastases from Colorectal Cancer. Ann. Surg. 2006, 244, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Mitry, E.; Guiu, B.; Cosconea, S.; Jooste, V.; Faivre, J.; Bouvier, A.-M. Epidemiology, management and prognosis of colorectal cancer with lung metastases: A 30-year population-based study. Gut 2010, 59, 1383–1388. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Du, L.; Yang, Y.; Wang, C.; Liu, H.; Wang, L.; Zhang, X.; Li, W.; Zheng, G.; Dong, Z. miR-429 is an independent prognostic factor in colorectal cancer and exerts its anti-apoptotic function by targeting SOX2. Cancer Lett. 2013, 329, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Leung, S.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008, 299, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Karlan, B.Y.; Oza, A.M.; Richardson, G.E.; Provencher, D.M.; Hansen, V.L.; Buck, M.; Chambers, S.K.; Ghatage, P.; Pippitt, C.H.; Brown, J.V.; et al. Randomized, double-blind, placebo-controlled phase II study of AMG 386 combined with weekly paclitaxel in patients with recurrent ovarian cancer. J. Clin. Oncol. 2012, 30, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Mita, A.C.; Takimoto, C.H.; Mita, M.; Tolcher, A.; Sankhala, K.; Sarantopoulos, J.; Valdivieso, M.; Wood, L.; Rasmussen, E.; Sun, Y.N.; et al. Phase 1 study of AMG 386, a selective angiopoietin 1/2-neutralizing peptibody, in combination with chemotherapy in adults with advanced solid tumors. Clin. Cancer Res. 2010, 16, 3044–3056. [Google Scholar] [CrossRef] [PubMed]
- Gattolliat, C.-H.; Uguen, A.; Pesson, M.; Trillet, K.; Simon, B.; Doucet, L.; Robaszkiewicz, M.; Corcos, L. MicroRNA and targeted mRNA expression profiling analysis in human colorectal adenomas and adenocarcinomas. Eur. J. Cancer 2015, 51, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Bandrés, E.; Cubedo, E.; Agirre, X.; Malumbres, R.; Zarate, R.; Ramirez, N.; Abajo, A.; Navarro, A.; Moreno, I.; Monzo, M.; et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 2006, 5, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, W.-J.; Yang, L.; Ma, Q.; Zhang, H.; Adell, G.; Arbman, G.; Wang, Z.-Q.; Li, Y.; Zhou, Z.-G.; Sun, X.-F. MicroRNA expression profile reveals miR-17-92 and miR-143-145 cluster in synchronous colorectal cancer. Medicine (Baltimore) 2015, 94. [Google Scholar] [CrossRef] [PubMed]
- Vychytilova-Faltejskova, P.; Pesta, M.; Radova, L.; Liska, V.; Daum, O.; Kala, Z.; Svoboda, M.; Kiss, I.; Slaby, O. Genome-wide microRNA Expression Profiling in Primary Tumors and Matched Liver Metastasis of Patients with Colorectal Cancer. Cancer Genom.-Proteom. 2016, 13, 311–316. [Google Scholar]
- Casazza, A.; di Conza, G.; Wenes, M.; Finisguerra, V.; Deschoemaeker, S.; Mazzone, M. Tumor stroma: A complexity dictated by the hypoxic tumor microenvironment. Oncogene 2014, 33, 1743–1754. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Lavotshkin, S.; Lyden, D. The secreted factors responsible for pre-metastatic niche formation: Old sayings and new thoughts. Semin. Cancer Biol. 2011, 21, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; de Sauvage, F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013, 501, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, S.; Zhang, J.D.; Schwäger, A.; Mannsperger, H.; Riazalhosseini, Y.; Burmester, S.; Ward, A.; Korf, U.; Wiemann, S.; Sahin, Ö. miR-200bc/429 cluster targets PLCγ1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene 2010, 29, 4297–4306. [Google Scholar] [CrossRef] [PubMed]
- Georges, S.A.; Biery, M.C.; Kim, S.; Schelter, J.M.; Guo, J.; Chang, A.N.; Jackson, A.L.; Carleton, M.O.; Linsley, P.S.; Cleary, M.A.; et al. Coordinated Regulation of Cell Cycle Transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008, 68, 10105–10112. [Google Scholar] [CrossRef] [PubMed]
- Khella, H.W.Z.; Bakhet, M.; Allo, G.; Jewett, M.A.S.; Girgis, A.H.; Latif, A.; Girgis, H.; Both, I.V.; Bjarnason, G.A.; Yousef, G.M. miR-192, miR-194 and miR-215: A convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis 2013, 34, 2231–2239. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Wang, Y.; Kudo, K.; Gavin, E.J.; Xi, Y.; Ju, J. miR-192 Regulates Dihydrofolate Reductase and Cellular Proliferation through the p53-microRNA Circuit. Clin. Cancer Res. 2008, 14, 8080–8086. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, P.; Hultine, S.; Smith, L.M.; Dews, M.; Fox, J.L.; Biyashev, D.; Schelter, J.M.; Huang, Q.; Cleary, M.A.; Volpert, O.V.; et al. p53-Responsive miR-194 Inhibits Thrombospondin-1 and Promotes Angiogenesis in Colon Cancers. Cancer Res. 2011, 71, 7490–7501. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Pecqueux, M.; Halama, N.; Dienemann, H.; Muley, T.; Pfannschmidt, J.; Lasitschka, F.; Klupp, F.; Schmidt, T.; Rahbari, N.; et al. Tumour-site-dependent expression profile of angiogenic factors in tumour-associated stroma of primary colorectal cancer and metastases. Br. J. Cancer 2013, 110. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Milazzo, M.; Chieco, P.; Negrini, M.; Calin, G.A.; Grazi, G.L.; Pollutri, D.; Croce, C.M.; Bolondi, L.; Gramantieri, L. miR-199a-3p Regulates mTOR and c-Met to Influence the Doxorubicin Sensitivity of Human Hepatocarcinoma Cells. Cancer Res. 2010, 70, 5184–5193. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, U.J.; Kim, M.N.; Lee, E.-J.; Kim, J.Y.; Lee, M.Y.; Choung, S.; Kim, Y.J.; Choi, Y.-C. MicroRNA miR-199a* Regulates the MET Proto-oncogene and the Downstream Extracellular Signal-regulated Kinase 2 (ERK2). J. Biol. Chem. 2008, 283, 18158–18166. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Jing, Y.; Li, W.; Qian, X.; Xu, Q.; Li, F.-S.; Liu, L.-Z.; Jiang, B.-H.; Jiang, Y. Roles and Mechanism of miR-199a and miR-125b in Tumor Angiogenesis. PLoS ONE 2013, 8, e56647. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.-Y.; Wang, Y.-X.; Yan, W.-T.; Li, H.-Y.; Tian, Y.-Z.; Wang, S.-M.; Zhao, H.-L. MicroRNA-125b Functions as a Tumor Suppressor in Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2012, 13, 8762–8774. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Zhang, L.; Meisgen, F.; Harada, M.; Heilborn, J.; Homey, B.; Grandér, D.; Ståhle, M.; Sonkoly, E.; Pivarcsi, A. MicroRNA-125b Down-regulates Matrix Metallopeptidase 13 and Inhibits Cutaneous Squamous Cell Carcinoma Cell Proliferation, Migration, and Invasion. J. Biol. Chem. 2012, 287, 29899–29908. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, M.; Zhu, S.; Wu, F.; Wu, H.; Walia, V.; Kumar, S.; Elble, R.; Watabe, K.; Mo, Y.-Y. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. USA 2009, 106, 3207–3212. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Xu, Q.; Mao, F.; Li, D.; Bian, C.; Liu, L.-Z.; Jiang, Y.; Chen, X.; Qi, Y.; Zhang, X.; et al. miR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle 2012, 11, 2137–2145. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Kanno, T.; Nakayamada, S.; Hirahara, K.; Sciumè, G.; Muljo, S.A.; Kuchen, S.; Casellas, R.; Wei, L.; Kanno, Y.; et al. TGF-β and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat. Immunol. 2012, 13, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.-H.; Li, H.; Wang, F.; Yu, J.; He, J.-S. The Tumor Suppressor Roles of miR-433 and miR-127 in Gastric Cancer. Int. J. Mol. Sci. 2013, 14, 14171–14184. [Google Scholar] [CrossRef] [PubMed]
- Pichiorri, F.; Suh, S.-S.; Rocci, A.; de Luca, L.; Taccioli, C.; Santhanam, R.; Wenchao, Z.; Benson, D.M.; Hofmainster, C.; Alder, H.; et al. Down-regulation of p53-inducible microRNAs 192, 194 and 215 impairs the p53/MDM2 auto-regulatory loop in multiple myeloma development. Cancer Cell 2010, 18, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Fu, X.; Chen, X.; Zeng, S.; Tian, Y.; Jove, R.; Xu, R.; Huang, W. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology 2010, 52, 2148–2157. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, T.; Fang, O.; Leach, L.J.; Hu, X.; Luo, Z. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27kip1. Oncogene 2014, 33, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, L.; Matyunina, L.V.; Hill, C.G.; McDonald, J.F. Overexpression of miR-429 induces mesenchymal-to-epithelial transition (MET) in metastatic ovarian cancer cells. Gynecol. Oncol. 2011, 121, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Van der Wal, G.E.; Gouw, A.S.; Kamps, J.A.; Moorlag, H.E.; Bulthuis, M.L.; Molema, G.; de Jong, K.P. Angiogenesis in synchronous and metachronous colorectal liver metastases: The liver as a permissive soil. Ann. Surg. 2012, 255, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Boni, V.; Bitarte, N.; Cristobal, I.; Zarate, R.; Rodriguez, J.; Maiello, E.; Garcia-Foncillas, J.; Bandres, E. miR-192/miR-215 Influence 5-Fluorouracil Resistance through Cell Cycle-Mediated Mechanisms Complementary to Its Post-transcriptional Thymidilate Synthase Regulation. Mol. Cancer Ther. 2010, 9, 2265–2275. [Google Scholar] [CrossRef] [PubMed]
- Mitchem, J.B.; Brennan, D.J.; Knolhoff, B.L.; Belt, B.A.; Zhu, Y.; Sanford, D.E.; Belaygorod, L.; Carpenter, D.; Collins, L.; Piwnica-Worms, D.; et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression and improves chemotherapeutic responses. Cancer Res. 2013, 73, 1128–1141. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Fiala, M.; Musso, G.; Halama, N.; Keim, S.; Mazzone, M.; Lasitschka, F.; Pecqueux, M.; Klupp, F.; Schmidt, T.; et al. Prognostic impact of a compartment-specific angiogenic marker profile in patients with pancreatic cancer. Oncotarget 2014, 5, 12978–12989. [Google Scholar] [CrossRef] [PubMed]
- Ibberson, D.; Benes, V.; Muckenthaler, M.U.; Castoldi, M. RNA degradation compromises the reliability of microRNA expression profiling. BMC Biotechnol. 2009, 9, 102. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Liver Metastases (n = 24) | Lung Metastases (n = 23) | |
|---|---|---|---|
| Age (years) | min–max | 40–76 | 36–79 |
| mean ± SD | 62 ± 10.32 | 64 ± 10.78 | |
| Sex | male | 16 | 15 |
| female | 8 | 8 | |
| Metastases | synchron. | 13 | 4 |
| metachron. | 11 | 19 | |
| PFS (months) | mean ± SD | 23.51 ± 26.18 | 68.60 ± 41.26 |
| median | 13.59 | 54.58 | |
| Events OS | total | 9 | 16 |
| % | 37.5 | 69.6 | |
| OS (months) | mean ± SD | 21.68 ± 15.26 | 24.11 ± 15.89 |
| Median | 16.51 | 22.75 | |
| Differential Expression in Liver and Lung Metastases | |||||||
|---|---|---|---|---|---|---|---|
| miRNA | Tissue | S vs. N | p-Value | T vs. N | p-Value | S vs. T | p-Value |
| miR-125b | liver | 3.417 | 0.438 | 0.070 | <0.0001 | 211.500 | <0.0001 |
| lung | 7.305 | 0.0078 | 0.206 | <0.0001 | 43.990 | <0.0001 | |
| miR-145 | liver | 4.048 | 0.533 | 0.095 | <0.0001 | 223.100 | <0.0001 |
| lung | 1.848 | 0.3465 | 0.203 | <0.0001 | 14.560 | <0.0001 | |
| miR-199a-3p | liver | 6.593 | 0.07 | 0.249 | <0.0001 | 624.400 | <0.0001 |
| lung | 4.700 | 0.0296 | 0.345 | <0.0001 | 14.480 | <0.0001 | |
| miR-199a-5p | liver | 9.137 | 0.0803 | 0.071 | <0.0001 | 510.900 | <0.0001 |
| lung | 9.143 | 0.0004 | 0.649 | 0.0016 | 25.560 | <0.0001 | |
| miR-429 | liver | 14.610 | 0.0065 | 46.150 | 0.0009 | 0.25 | 0.0066 |
| lung | 9.418 | 0.0163 | 80.890 | <0.0001 | 0.178 | <0.0001 | |
| miR-127-3p | liver | 1.310 | 0.0658 | 0.101 | <0.0001 | 61.180 | <0.0001 |
| lung | 4.907 | 0.0149 | 0.702 | 0.0023 | 10.950 | <0.0001 | |
| miR-19b | liver | 0.278 | <0.0001 | 0.291 | <0.0001 | 2.569 | 0.2699 |
| lung | 1.944 | 0.0006 | 2.326 | 0.0007 | 1.475 | 0.0383 | |
| miR-194 | liver | 0.349 | <0.0001 | 3.103 | 0.7265 | 0.466 | <0.0001 |
| lung | 168.500 | <0.0001 | 714.900 | <0.0001 | 0.272 | <0.0001 | |
| miR-215b | liver | 0.167 | <0.0001 | 1.147 | 0.0372 | 0.392 | <0.0001 |
| lung | 58.310 | 0.0028 | 319.100 | <0.0001 | 0.095 | <0.0001 | |
| miR-192 | liver | 0.030 | <0.0001 | 0.306 | <0.0001 | 0,255 | <0.0001 |
| lung | 33.480 | 0.0012 | 192.100 | <0.0001 | 0,198 | <0.0001 | |
| miR-21 | liver | 13.900 | 0.0006 | 2.872 | 0.2296 | 33.680 | <0.0001 |
| lung | 11.880 | 0.0001 | 5.653 | 0.0025 | 4.557 | 0.0045 | |
| miRNA | Tissue | FC Liver vs. Lung (Mean) | SEM | p-Value |
|---|---|---|---|---|
| miR-125 | Stroma | 1.00 | 0.14 | 0.80 |
| Tumor | −2.5 | 1.01 | 0.003 | |
| Host tissue | 1.86 | 1.82 | 0.013 | |
| miR-145 | Stroma | 2.33 | 0.79 | 0.54 |
| Tumor | −5.5 | 0.22 | 0.0002 | |
| Host tissue | −1.2 | 0.49 | 0.51 | |
| miR-199-3 | Stroma | 1.32 | 1.04 | 0.72 |
| Tumor | −5.00 | 0.16 | <0.0001 | |
| Host tissue | 1.06 | 1.68 | 0.73 | |
| miR-199-5 | Stroma | 1.32 | 0.47 | 0.85 |
| Tumor | −12.5 | 0.14 | <0.0001 | |
| Host tissue | 2.85 | 2.64 | 0.008 | |
| miR-429 | Stroma | −1.56 | 0.30 | 0.26 |
| Tumor | −5.56 | 0.14 | 0.0002 | |
| Host tissue | −2.22 | 2.47 | 0.017 | |
| miR-19 | Stroma | −1.56 | 0.30 | 0.04 |
| Tumor | −3.57 | 0.19 | 0.0001 | |
| Host tissue | 3.70 | 3.38 | 0.0001 | |
| miR-127 | Stroma | 1.05 | 0.56 | 0.91 |
| Tumor | −2.38 | 0.25 | 0.02 | |
| Host tissue | 4.18 | 3.13 | <0.0001 | |
| miR-194 | Stroma | −2.63 | 0.46 | 0.03 |
| Tumor | −1.59 | 0.17 | 0.03 | |
| Host tissue | 156 | 1742 | <0.0001 | |
| miR-215 | Stroma | −1.72 | 0.37 | 0.01 |
| Tumor | −3.33 | 0.21 | 0.0004 | |
| Host tissue | 108 | 83.25 | <0.0001 | |
| miR-192 | Stroma | −3.33 | 0.18 | <0.0001 |
| Tumor | −2.71 | 0.18 | 0.001 | |
| Host tissue | 347 | 598 | <0.0001 | |
| miR-21 | Stroma | −1.92 | 0.21 | 0.04 |
| Tumor | −7.14 | 0.17 | <0.0001 | |
| Host tissue | −1.89 | 2.30 | 0.18 |
| miRNA | Liver | Lung | Liver and Lung | ||||||
|---|---|---|---|---|---|---|---|---|---|
| <median | >median | p | <median | >median | p | <median | >median | p | |
| miR-125ds | 34.9 | 46.2 | 0.25 | 37.9 | 31.4 | 0.57 | 36.6 | 39.3 | 0.70 |
| miR-125dt | 36.0 | 38.8 | 0.28 | 30.9 | 36.9 | 0.82 | 33.8 | 43.7 | 0.40 |
| miR-125dn | 27.2 | 56.9 | 0.05 | 32.8 | 37.0 | 0.71 | 32.5 | 45.3 | 0.12 |
| miR-145ds | 41.3 | 43.1 | 0.84 | 36.6 | 34.1 | 0.67 | 37.9 | 39.3 | 0.98 |
| miR-145dt | 46.0 | 31.2 | 0.46 | 36.3 | 33.4 | 0.74 | 41.0 | 34.6 | 0.36 |
| miR-145dn | 25.3 | 55.9 | 0.005 | 33.5 | 34.7 | 0.75 | 31.4 | 45.6 | 0.038 |
| miR-199-3ds | 27.9 | 54.7 | 0.05 | 35.6 | 35.3 | 0.76 | 33.0 | 45.3 | 0.23 |
| miR-199-3dt | 43.2 | 32.8 | 0.98 | 32.2 | 38.6 | 0.69 | 37.0 | 41.4 | 0.79 |
| miR-199-3dn | 28.9 | 51.1 | 0.08 | 35.1 | 34.5 | 0.76 | 33.0 | 44.2 | 0.42 |
| miR-199-5ds | 41.3 | 43.1 | 0.84 | 36.0 | 33.8 | 0.67 | 38.5 | 38.8 | 0.85 |
| miR-199-5dt | 41.5 | 33.6 | 0.81 | 35.6 | 33.5 | 0.62 | 38.4 | 39.0 | 0.87 |
| miR-199-5dn | 25.9 | 55.9 | 0.008 | 41.2 | 30.4 | 0.39 | 35.3 | 43.0 | 0.28 |
| miR-429ds | 40.9 | 41.7 | 0.83 | 37.4 | 32.4 | 0.65 | 39.6 | 36.7 | 0.88 |
| miR-429dt | 45.2 | 31.5 | 0.56 | 35.9 | 35.0 | 0.98 | 40.6 | 36.7 | 0.73 |
| miR-429dn | 27.4 | 55.4 | 0.03 | 30.6 | 41.2 | 0.43 | 31.1 | 49.3 | 0.06 |
| miR-19ds | 42.6 | 42.8 | 0.97 | 32.5 | 36.1 | 0.69 | 37.1 | 39.5 | 0.77 |
| miR-19dt | 45.0 | 32.0 | 0.59 | 39.4 | 31.1 | 0.54 | 43.8 | 32.4 | 0.33 |
| miR-19dn | 28.9 | 51.1 | 0.12 | 34.1 | 36.7 | 0.75 | 34.2 | 43.6 | 0.21 |
| miR-127ds | 35.2 | 46.7 | 0.48 | 32.0 | 38.1 | 0.59 | 33.1 | 44.3 | 0.32 |
| miR-127dt | 44.5 | 33.1 | 0.68 | 38.5 | 27.4 | 0.30 | 42.3 | 30.6 | 0.34 |
| miR-127dn | 22.3 | 57.1 | 0.001 | 38.8 | 31.7 | 0.62 | 33.7 | 44.1 | 0.12 |
| miR-194ds | 42.1 | 41.0 | 0.97 | 35.0 | 34.2 | 0.96 | 39.0 | 37.5 | 0.88 |
| miR-194dt | 39.6 | 36.1 | 0.52 | 28.2 | 34.3 | 0.18 | 33.1 | 46.8 | 0.16 |
| miR-194dn | 24.5 | 56.1 | 0.003 | 40.6 | 27.0 | 0.42 | 34.7 | 42.4 | 0.19 |
| miR-215ds | 41.8 | 38.3 | 0.68 | 37.9 | 30.9 | 0.52 | 41.0 | 35.3 | 0.39 |
| miR-215dt | 47.6 | 37.7 | 0.48 | 36.7 | 31.9 | 0.56 | 40.8 | 36.4 | 0.44 |
| miR-215dn | 19.7 | 57.1 | 0.000 | 34.3 | 34.3 | 0.85 | 23.4 | 47.8 | 0.04 |
| miR-192ds | 43.5 | 39.5 | 0.72 | 35.1 | 34.3 | 0.99 | 40.5 | 37.8 | 0.76 |
| miR-192dt | 46.0 | 35.1 | 0.88 | 29.9 | 39.9 | 0.44 | 36.0 | 41.3 | 0.69 |
| miR-192dn | 26.0 | 55.9 | 0.015 | 44.4 | 24.9 | 0.13 | 37.8 | 40.3 | 0.58 |
| miR-21ds | 44.1 | 33.1 | 0.56 | 38.4 | 29.0 | 0.45 | 42.2 | 31.8 | 0.42 |
| miR-21dt | 52.0 | 31.8 | 0.29 | 28.4 | 41.3 | 0.33 | 36.5 | 39.7 | 0.96 |
| miR-21dn | 31.8 | 50.2 | 0.20 | 40.7 | 27.0 | 0.35 | 38.9 | 39.4 | 0.83 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pecqueux, M.; Liebetrau, I.; Werft, W.; Dienemann, H.; Muley, T.; Pfannschmidt, J.; Müssle, B.; Rahbari, N.; Schölch, S.; Büchler, M.W.; et al. A Comprehensive MicroRNA Expression Profile of Liver and Lung Metastases of Colorectal Cancer with Their Corresponding Host Tissue and Its Prognostic Impact on Survival. Int. J. Mol. Sci. 2016, 17, 1755. https://doi.org/10.3390/ijms17101755
Pecqueux M, Liebetrau I, Werft W, Dienemann H, Muley T, Pfannschmidt J, Müssle B, Rahbari N, Schölch S, Büchler MW, et al. A Comprehensive MicroRNA Expression Profile of Liver and Lung Metastases of Colorectal Cancer with Their Corresponding Host Tissue and Its Prognostic Impact on Survival. International Journal of Molecular Sciences. 2016; 17(10):1755. https://doi.org/10.3390/ijms17101755
Chicago/Turabian StylePecqueux, Mathieu, Isabell Liebetrau, Wiebke Werft, Hendrik Dienemann, Thomas Muley, Joachim Pfannschmidt, Benjamin Müssle, Nuh Rahbari, Sebastian Schölch, Markus W. Büchler, and et al. 2016. "A Comprehensive MicroRNA Expression Profile of Liver and Lung Metastases of Colorectal Cancer with Their Corresponding Host Tissue and Its Prognostic Impact on Survival" International Journal of Molecular Sciences 17, no. 10: 1755. https://doi.org/10.3390/ijms17101755






