Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack
Abstract
:1. Introduction
2. CTC: From Quantity to Quality
2.1. CTC: The Premise
2.2. CTC as Surrogate Biomarkers
2.3. CTC: Open Questions10.3390/ijms17101775
3. CTC Intrinsic Heterogeneity
3.1. Biophysical Features: Small Size CTC and Clusters
3.2. Identification of CTC Subsets with Specific Properties
3.2.1. CTC in Epithelial-to-Mesenchymal Transition
3.2.2. CTC with Stem Cell Properties
3.2.3. CTC Undergoing Metabolic Reprogramming
3.2.4. CTC with Metastasis-Initiating Properties
3.2.5. Apoptotic CTC
3.2.6. Drug-Targetable CTC
3.3. Genomic Heterogeneity
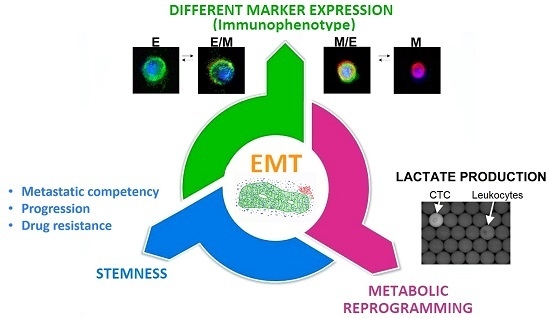
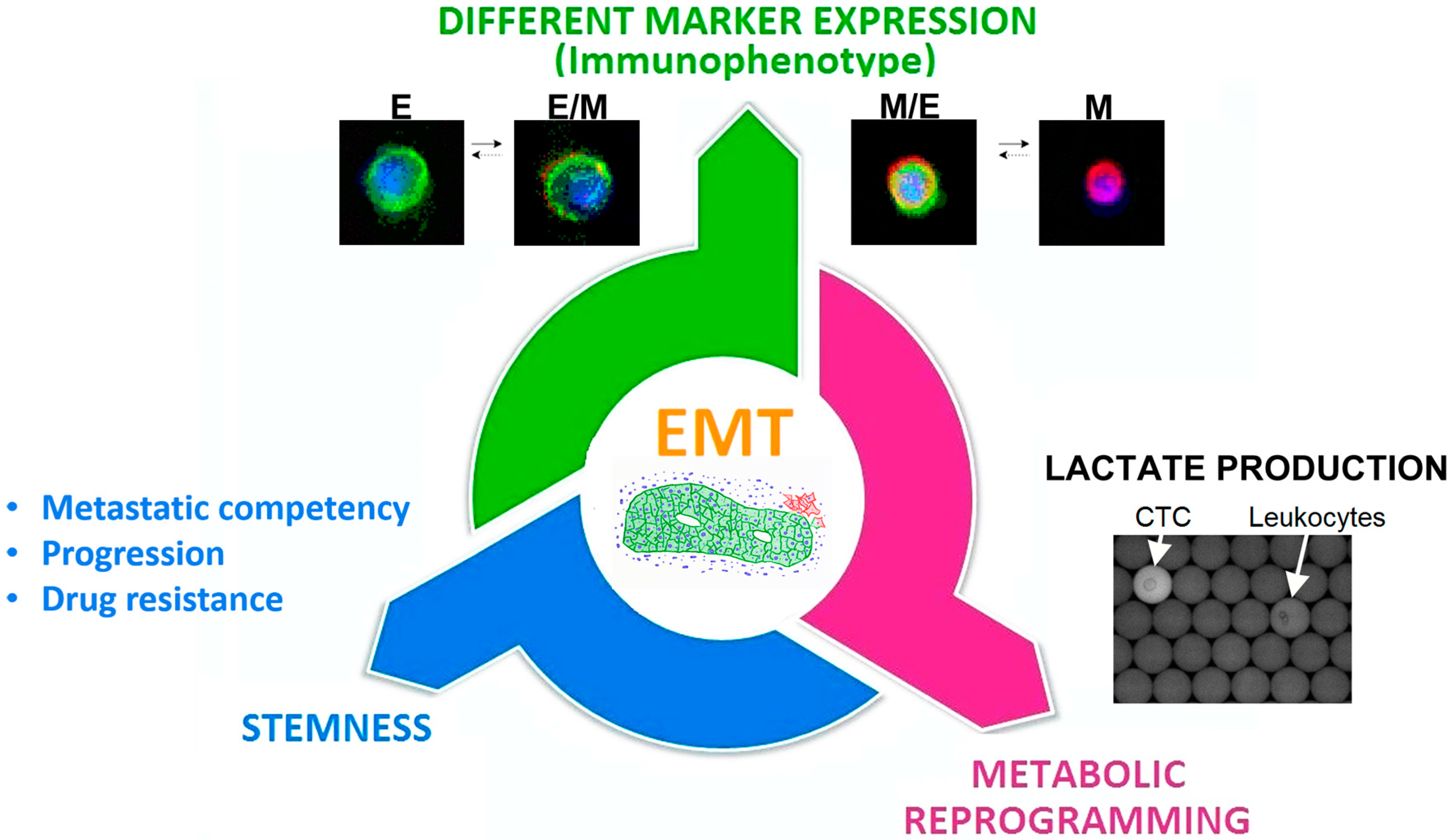
3.4. The Epithelial-to-Mesenchymal Transition as a Common Trigger of Different Types of Tumor Heterogeneity
4. Summary and Conclusions
5. Methods
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ALDH1 | Aldehyde dehydrogenase 1 |
| cfDNA | Circulating free DNA |
| CSC | Cancer Stem Cells |
| CTC | Circulating Tumor Cells |
| EMT | Epithelial-to-Mesenchymal Transition |
| ER | Estrogen Receptor |
| EpCAM | Epithelial Cell Adhesion Molecule |
| EGFR | Epidermal Growth Factor Receptor |
| HER2 | Human epidermal growth factor receptor 2 |
| MBC | Metastatic Breast Cancer |
| NGS | Next Generation Sequencing |
| OS | Overall Survival |
| PgR | Progesteron Receptor |
| PIK3 | Phosphatidylinositol 3-kinase |
| PIK3CA | Catalytic subunit of PIK3 |
| PFS | Progression Free Survival |
| SCS | Single Cell Sequencing |
References
- Siu, L.L. Accounting for tumor heterogeneity in the development of predictive biomarkers. Clin. Adv. Hematol. Oncol. 2013, 11, 312–313. [Google Scholar] [PubMed]
- Gagan, J.; van Allen, E.M. Next-generation sequencing to guide cancer therapy. Genome Med. 2015, 7, 80. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Paik, S. Gene-expression-based prognostic assays for breast cancer. Nat. Rev. Clin. Oncol. 2010, 7, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Speicher, M.R.; Pantel, K. Tumor signatures in the blood. Nat. Biotechnol. 2014, 32, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bauerle, T.; Wallwiener, M.; et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013, 31, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 2013, 5, 180ra48. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Makarovskiy, A.N.; Ackerley, W., 3rd; Wojcik, L.; Halpert, G.K.; Stein, B.S.; Carreiro, M.P.; Hixson, D.C. Application of immunomagnetic beads in combination with RT-PCR for the detection of circulating prostate cancer cells. J. Clin. Lab. Anal. 1997, 11, 346–350. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.; Uhr, J.W.; Terstappen, L.W. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Naoe, M.; Ogawa, Y.; Morita, J.; Omori, K.; Takeshita, K.; Shichijyo, T.; Okumura, T.; Igarashi, A.; Yanaihara, A.; Iwamoto, S.; et al. Detection of circulating urothelial cancer cells in the blood using the CellSearch System. Cancer 2007, 109, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Wolfrum, F.; Vogel, I.; Fandrich, F.; Kalthoff, H. Detection and clinical implications of minimal residual disease in gastro-intestinal cancer. Langenbecks Arch. Surg. 2005, 390, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Choy, A.; McCulloch, P.G. Detection and enumeration of circulating tumour cells in colorectal cancer. Br. J. Surg. 1993, 80, 1490. [Google Scholar] [CrossRef] [PubMed]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Maheswaran, S.; Haber, D.A. Circulating tumor cells: A window into cancer biology and metastasis. Curr. Opin. Genet. Dev. 2010, 20, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Yeatman, T.J.; Nicolson, G.L. Molecular basis of tumor progression: Mechanisms of organ-specific tumor metastasis. Semin. Surg. Oncol. 1993, 9, 256–263. [Google Scholar] [PubMed]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Oskarsson, T.; Acharyya, S.; Nguyen, D.X.; Zhang, X.H.; Norton, L.; Massague, J. Tumor self-seeding by circulating cancer cells. Cell 2009, 139, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. EMT and dissemination precede pancreatic tumor formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Thege, F.I.; Santana, S.M.; Lannin, T.B.; Saha, T.N.; Tsai, S.; Maggs, L.R.; Kochman, M.L.; Ginsberg, G.G.; Lieb, J.G.; et al. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology 2014, 146, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Hofman, V.; Long-Mira, E.; Selva, E.; Vignaud, J.M.; Padovani, B.; Mouroux, J.; Marquette, C.H.; Hofman, P. “Sentinel” circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS ONE 2014, 9, e111597. [Google Scholar] [CrossRef] [PubMed]
- Lianidou, E.S.; Mavroudis, D.; Georgoulias, V. Clinical challenges in the molecular characterization of circulating tumour cells in breast cancer. Br. J. Cancer 2013, 108, 2426–2432. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Hou, J.M.; Ward, T.H.; Blackhall, F.H.; Dive, C. Circulating tumour cells: Their utility in cancer management and predicting outcomes. Ther. Adv. Med. Oncol. 2010, 2, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.C.; Doyle, G.V.; Terstappen, L.W. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J. Oncol. 2010, 2010, 617421. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Fritsche, H.; Muller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Janicke, F.; et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the CellSearch system. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Riethdorf, S.; Wu, G.; Wang, T.; Yang, K.; Peng, G.; Liu, J.; Pantel, K. Meta-analysis of the prognostic value of circulating tumor cells in breast cancer. Clin. Cancer Res. 2012, 18, 5701–5710. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.C.; Peeters, D.J.; Fehm, T.; Nole, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Schramm, A.; Friedl, T.W.; Schochter, F.; Scholz, C.; de Gregorio, N.; Huober, J.; Rack, B.; Trapp, E.; Alunni-Fabbroni, M.; Muller, V.; et al. Therapeutic intervention based on circulating tumor cell phenotype in metastatic breast cancer: Concept of the DETECT study program. Arch. Gynecol. Obstet. 2016, 293, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Smerage, J.B.; Barlow, W.E.; Hortobagyi, G.N.; Winer, E.P.; Leyland-Jones, B.; Srkalovic, G.; Tejwani, S.; Schott, A.F.; O’Rourke, M.A.; Lew, D.L.; et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J. Clin. Oncol. 2014, 32, 3483–3489. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.C.; Fehm, T.; Ignatiadis, M.; Smerage, J.B.; Alix-Panabieres, C.; Janni, W.; Messina, C.; Paoletti, C.; Muller, V.; Hayes, D.F.; et al. Clinical application of circulating tumor cells in breast cancer: Overview of the current interventional trials. Cancer Metastasis Rev. 2013, 32, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224. [Google Scholar] [CrossRef] [PubMed]
- Lucci, A.; Hall, C.S.; Lodhi, A.K.; Bhattacharyya, A.; Anderson, A.E.; Xiao, L.; Bedrosian, I.; Kuerer, H.M.; Krishnamurthy, S. Circulating tumour cells in non-metastatic breast cancer: A prospective study. Lancet Oncol. 2012, 13, 688–695. [Google Scholar] [CrossRef]
- Pierga, J.Y.; Hajage, D.; Bachelot, T.; Delaloge, S.; Brain, E.; Campone, M.; Dieras, V.; Rolland, E.; Mignot, L.; Mathiot, C.; et al. High independent prognostic and predictive value of circulating tumor cells compared with serum tumor markers in a large prospective trial in first-line chemotherapy for metastatic breast cancer patients. Ann. Oncol. 2012, 23, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Basso, U.; Celadin, R.; Zilio, F.; Pucciarelli, S.; Aieta, M.; Barile, C.; Sava, T.; Bonciarelli, G.; Tumolo, S.; et al. M30 neoepitope expression in epithelial cancer: Quantification of apoptosis in circulating tumor cells by CellSearch analysis. Clin. Cancer Res. 2010, 16, 5233–5243. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Rothe, F.; Chaboteaux, C.; Durbecq, V.; Rouas, G.; Criscitiello, C.; Metallo, J.; Kheddoumi, N.; Singhal, S.K.; Michiels, S.; et al. HER2-positive circulating tumor cells in breast cancer. PLoS ONE 2011, 6, e15624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Guan, Y.; Sun, Y.; Ai, D.; Guo, Q. Tumor heterogeneity and circulating tumor cells. Cancer Lett. 2016, 374, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Nakamura, S.; Tokuda, Y.; Shimoda, Y.; Yagata, H.; Yoshida, A.; Ota, H.; Hortobagyi, G.N.; Cristofanilli, M.; Ueno, N.T. Prognostic value of HER2-positive circulating tumor cells in patients with metastatic breast cancer. Int. J. Clin. Oncol. 2012, 17, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Pestrin, M.; Bessi, S.; Galardi, F.; Truglia, M.; Biggeri, A.; Biagioni, C.; Cappadona, S.; Biganzoli, L.; Giannini, A.; Di Leo, A. Correlation of HER2 status between primary tumors and corresponding circulating tumor cells in advanced breast cancer patients. Breast Cancer Res. Treat. 2009, 118, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Fehm, T.; Muller, V.; Aktas, B.; Janni, W.; Schneeweiss, A.; Stickeler, E.; Lattrich, C.; Lohberg, C.R.; Solomayer, E.; Rack, B.; et al. HER2 status of circulating tumor cells in patients with metastatic breast cancer: A prospective, multicenter trial. Breast Cancer Res. Treat. 2010, 124, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009, 11, R46. [Google Scholar] [CrossRef] [PubMed]
- Tewes, M.; Aktas, B.; Welt, A.; Mueller, S.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: An option for monitoring response to breast cancer related therapies. Breast Cancer Res. Treat. 2009, 115, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, S.V.; Bingham, C.; Fittipaldi, P.; Austin, L.; Palazzo, J.; Palmer, G.; Alpaugh, K.; Cristofanilli, M. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breast cancer patients. Breast Cancer Res. 2014, 16, 445. [Google Scholar] [CrossRef] [PubMed]
- De Luca, F.; Rotunno, G.; Salvianti, F.; Galardi, F.; Pestrin, M.; Gabellini, S.; Simi, L.; Mancini, I.; Vannucchi, A.M.; Pazzagli, M.; et al. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget 2016, 7, 26107–26119. [Google Scholar] [PubMed]
- Bulfoni, M.; Gerratana, L.; Del Ben, F.; Marzinotto, S.; Sorrentino, M.; Turetta, M.; Scoles, G.; Toffoletto, B.; Isola, M.; Beltrami, C.A.; et al. In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-to-mesenchymal transition is associated with a poor prognosis. Breast Cancer Res. 2016, 18, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pestrin, M.; Salvianti, F.; Galardi, F.; de Luca, F.; Turner, N.; Malorni, L.; Pazzagli, M.; di Leo, A.; Pinzani, P. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol. Oncol. 2015, 9, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Polzer, B.; Medoro, G.; Pasch, S.; Fontana, F.; Zorzino, L.; Pestka, A.; Andergassen, U.; Meier-Stiegen, F.; Czyz, Z.T.; Alberter, B.; et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol. Med. 2014, 6, 1371–1386. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Tripathy, D.; Shete, S.; Ashfaq, R.; Haley, B.; Perkins, S.; Beitsch, P.; Khan, A.; Euhus, D.; Osborne, C.; et al. HER-2 gene amplification can be acquired as breast cancer progresses. Proc. Natl. Acad. Sci. USA 2004, 101, 9393–9398. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.M.; Fehm, T.; Orsini, M.; Cayrefourcq, L.; Maudelonde, T.; Pantel, K.; Alix-Panabieres, C. Prognostic relevance of viable circulating tumor cells detected by EPISPOT in metastatic breast cancer patients. Clin. Chem. 2014, 60, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Farace, F.; Massard, C.; Vimond, N.; Drusch, F.; Jacques, N.; Billiot, F.; Laplanche, A.; Chauchereau, A.; Lacroix, L.; Planchard, D.; et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br. J. Cancer 2011, 105, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Stathopoulou, A.; Gizi, A.; Perraki, M.; Apostolaki, S.; Malamos, N.; Mavroudis, D.; Georgoulias, V.; Lianidou, E.S. Real-time quantification of CK-19 mRNA-positive cells in peripheral blood of breast cancer patients using the lightcycler system. Clin. Cancer Res. 2003, 9, 5145–5151. [Google Scholar] [PubMed]
- Giordano, A.; Gao, H.; Anfossi, S.; Cohen, E.; Mego, M.; Lee, B.N.; Tin, S.; De Laurentiis, M.; Parker, C.A.; Alvarez, R.H.; et al. Epithelial-mesenchymal transition and stem cell markers in patients with HER2-positive metastatic breast cancer. Mol. Cancer Ther. 2012, 11, 2526–2534. [Google Scholar] [CrossRef] [PubMed]
- Barriere, G.; Riouallon, A.; Renaudie, J.; Tartary, M.; Rigaud, M. Mesenchymal and stemness circulating tumor cells in early breast cancer diagnosis. BMC Cancer 2012, 12, 114. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, N.; Meier-Stiegen, F.; Banys, M.; Neubauer, H.; Ruckhaeberle, E.; Fehm, T. Expression of stem cell and epithelial-mesenchymal transition markers in circulating tumor cells of breast cancer patients. BioMed Res. Int. 2014, 2014, 415721. [Google Scholar] [CrossRef] [PubMed]
- May, C.D.; Sphyris, N.; Evans, K.W.; Werden, S.J.; Guo, W.; Mani, S.A. Epithelial-mesenchymal transition and cancer stem cells: A dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, C.; Gradilone, A.; Naso, G.; Vincenzi, B.; Petracca, A.; Nicolazzo, C.; Palazzo, A.; Saltarelli, R.; Spremberg, F.; Cortesi, E.; et al. Epithelial-mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Res. Treat. 2011, 130, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Rose, L.; Zollner, T.M.; Krahn, T.; von Ahsen, O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Lustberg, M.B.; Balasubramanian, P.; Miller, B.; Garcia-Villa, A.; Deighan, C.; Wu, Y.; Carothers, S.; Berger, M.; Ramaswamy, B.; Macrae, E.R.; et al. Heterogeneous atypical cell populations are present in blood of metastatic breast cancer patients. Breast Cancer Res. 2014, 16, R23. [Google Scholar] [CrossRef] [PubMed]
- Sieuwerts, A.M.; Kraan, J.; Bolt, J.; van der Spoel, P.; Elstrodt, F.; Schutte, M.; Martens, J.W.; Gratama, J.W.; Sleijfer, S.; Foekens, J.A. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J. Natl. Cancer Inst. 2009, 101, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Blick, T.; Widodo, E.; Hugo, H.; Waltham, M.; Lenburg, M.E.; Neve, R.M.; Thompson, E.W. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin. Exp. Metastasis 2008, 25, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Bartkowiak, K.; Pantel, K. Functional studies on circulating and disseminated tumor cells in carcinoma patients. Mol. Oncol. 2016, 10, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Danila, D.C.; Pantel, K.; Fleisher, M.; Scher, H.I. Circulating tumors cells as biomarkers: Progress toward biomarker qualification. Cancer J. 2011, 17, 438–450. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Ho, H.; Lichterman, J.; Lu, Y.T.; Zhang, Y.; Garcia, M.A.; Chen, S.F.; Liang, A.J.; Hodara, E.; Zhau, H.E.; et al. Subclassification of prostate cancer circulating tumor cells by nuclear size reveals very small nuclear circulating tumor cells in patients with visceral metastases. Cancer 2015, 121, 3240–3251. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, A.S.; Ferraldeschi, R.; Krupa, R.; Landers, M.; Graf, R.; Louw, J.; Jendrisak, A.; Bales, N.; Marrinucci, D.; Zafeiriou, Z.; et al. Phenotypic diversity of circulating tumour cells in patients with metastatic castration-resistant prostate cancer. BJU Int. 2016. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Boareto, M.; Huang, B.; Jia, D.; Lu, M.; Ben-Jacob, E.; Onuchic, J.N.; Levine, H. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front. Oncol. 2015, 5, 155. [Google Scholar] [CrossRef] [PubMed]
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.H.; Wendel, M.; Luttgen, M.; Yoshioka, C.; Marrinucci, D.; Lazar, D.; Schram, E.; Nieva, J.; Bazhenova, L.; Morgan, A.; et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys. Biol. 2012, 9, 016001. [Google Scholar] [CrossRef] [PubMed]
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 2015, 12, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Forte, V.A.; Barrak, D.K.; Elhodaky, M.; Tung, L.; Snow, A.; Lang, J.E. The potential for liquid biopsies in the precision medical treatment of breast cancer. Cancer Biol. Med. 2016, 13, 19–40. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Kasimir-Bauer, S.; Hoffmann, O.; Wallwiener, D.; Kimmig, R.; Fehm, T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012, 14, R15. [Google Scholar] [CrossRef] [PubMed]
- Mego, M.; Gao, H.; Lee, B.N.; Cohen, E.N.; Tin, S.; Giordano, A.; Wu, Q.; Liu, P.; Nieto, Y.; Champlin, R.E.; et al. Prognostic Value of EMT-Circulating Tumor Cells in Metastatic Breast Cancer Patients Undergoing High-Dose Chemotherapy with Autologous Hematopoietic Stem Cell Transplantation. J. Cancer 2012, 3, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Mego, M.; De Giorgi, U.; Dawood, S.; Wang, X.; Valero, V.; Andreopoulou, E.; Handy, B.; Ueno, N.T.; Reuben, J.M.; Cristofanilli, M. Characterization of metastatic breast cancer patients with nondetectable circulating tumor cells. Int. J. Cancer 2011, 129, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, A.J.; Marengo, M.S.; Oltean, S.; Kemeny, G.; Bitting, R.L.; Turnbull, J.D.; Herold, C.I.; Marcom, P.K.; George, D.J.; Garcia-Blanco, M.A. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol. Cancer Res. 2011, 9, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Papadaki, M.A.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011, 13, R59. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Konstantinidis, G.; Markomanolaki, H.; Papadaki, M.A.; Mavroudis, D.; Stournaras, C.; Georgoulias, V.; Agelaki, S. Apoptotic circulating tumor cells in early and metastatic breast cancer patients. Mol. Cancer Ther. 2013, 12, 1886–1895. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulos, P.A.; Polioudaki, H.; Agelaki, S.; Kallergi, G.; Saridaki, Z.; Mavroudis, D.; Georgoulias, V. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010, 288, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Morel, A.P.; Lievre, M.; Thomas, C.; Hinkal, G.; Ansieau, S.; Puisieux, A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE 2008, 3, e2888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, S.; Liu, Y.; Liu, J.; Ma, Y.; Zhu, Y.; Zhang, J. Effects of the combination of RAD001 and docetaxel on breast cancer stem cells. Eur. J. Cancer 2012, 48, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- Bachelot, T.; Bourgier, C.; Cropet, C.; Ray-Coquard, I.; Ferrero, J.M.; Freyer, G.; Abadie-Lacourtoisie, S.; Eymard, J.C.; Debled, M.; Spaeth, D.; et al. Randomized phase II trial of everolimus in combination with tamoxifen in patients with hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer with prior exposure to aromatase inhibitors: A GINECO study. J. Clin. Oncol. 2012, 30, 2718–2724. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Campone, M.; Piccart, M.; Burris, H.A., 3rd; Rugo, H.S.; Sahmoud, T.; Noguchi, S.; Gnant, M.; Pritchard, K.I.; Lebrun, F.; et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 2012, 366, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Hurvitz, S.A.; Andre, F.; Jiang, Z.; Shao, Z.; Mano, M.S.; Neciosup, S.P.; Tseng, L.M.; Zhang, Q.; Shen, K.; Liu, D.; et al. Combination of everolimus with trastuzumab plus paclitaxel as first-line treatment for patients with HER2-positive advanced breast cancer (BOLERO-1): A phase 3, randomised, double-blind, multicentre trial. Lancet Oncol. 2015, 16, 816–829. [Google Scholar] [CrossRef]
- Andre, F.; O’Regan, R.; Ozguroglu, M.; Toi, M.; Xu, B.; Jerusalem, G.; Masuda, N.; Wilks, S.; Arena, F.; Isaacs, C.; et al. Everolimus for women with trastuzumab-resistant, HER2-positive, advanced breast cancer (BOLERO-3): A randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 2014, 15, 580–591. [Google Scholar] [CrossRef]
- Du, B.; Shim, J.S. Targeting Epithelial-Mesenchymal Transition (EMT) to Overcome Drug Resistance in Cancer. Molecules 2016, 21, 7. [Google Scholar] [CrossRef] [PubMed]
- Ithimakin, S.; Day, K.C.; Malik, F.; Zen, Q.; Dawsey, S.J.; Bersano-Begey, T.F.; Quraishi, A.A.; Ignatoski, K.W.; Daignault, S.; Davis, A.; et al. HER2 drives luminal breast cancer stem cells in the absence of HER2 amplification: Implications for efficacy of adjuvant trastuzumab. Cancer Res. 2013, 73, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Kim, G.I.; Davis, A.; Malik, F.; Henry, N.L.; Ithimakin, S.; Quraishi, A.A.; Tawakkol, N.; D’Angelo, R.; Paulson, A.K.; et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol. Cell. 2012, 47, 570–584. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Del Ben, F.; Turetta, M.; Celetti, G.; Piruska, A.; Bulfoni, M.; Cesselli, D.; Huck, W.T.; Scoles, G. A Method for Detecting Circulating Tumor Cells Based on the Measurement of Single-Cell Metabolism in Droplet-Based Microfluidics. Angew. Chem. Int. Ed. Engl. 2016, 55, 8581–8584. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Rugge, M.; Facchinetti, A.; Pizzi, M.; Nardo, G.; Barbieri, V.; Manicone, M.; De Faveri, S.; Chiara Scaini, M.; Basso, U.; et al. Retaining the long-survive capacity of Circulating Tumor Cells (CTCs) followed by xeno-transplantation: Not only from metastatic cancer of the breast but also of prostate cancer patients. Oncoscience 2014, 1, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Mehes, G.; Witt, A.; Kubista, E.; Ambros, P.F. Circulating breast cancer cells are frequently apoptotic. Am. J. Pathol. 2001, 159, 17–20. [Google Scholar] [CrossRef]
- Rossi, E.; Fassan, M.; Aieta, M.; Zilio, F.; Celadin, R.; Borin, M.; Grassi, A.; Troiani, L.; Basso, U.; Barile, C.; et al. Dynamic changes of live/apoptotic circulating tumour cells as predictive marker of response to sunitinib in metastatic renal cancer. Br. J. Cancer 2012, 107, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Aktas, B.; Kasimir-Bauer, S.; Muller, V.; Janni, W.; Fehm, T.; Wallwiener, D.; Pantel, K.; Tewes, M.; DETECT Study Group. Comparison of the HER2, estrogen and progesterone receptor expression profile of primary tumor, metastases and circulating tumor cells in metastatic breast cancer patients. BMC Cancer 2016, 16, 522. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Muller, V.; Zhang, L.; Rau, T.; Loibl, S.; Komor, M.; Roller, M.; Huober, J.; Fehm, T.; Schrader, I.; et al. Detection and HER2 expression of circulating tumor cells: Prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin. Cancer Res. 2010, 16, 2634–2645. [Google Scholar] [CrossRef] [PubMed]
- Berns, K.; Horlings, H.M.; Hennessy, B.T.; Madiredjo, M.; Hijmans, E.M.; Beelen, K.; Linn, S.C.; Gonzalez-Angulo, A.M.; Stemke-Hale, K.; Hauptmann, M.; et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007, 12, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, P.J.; Gili, M.; Scaltriti, M.; Serra, V.; Guzman, M.; Nijkamp, W.; Beijersbergen, R.L.; Valero, V.; Seoane, J.; Bernards, R.; et al. Phosphatidylinositol 3-kinase hyperactivation results in lapatinib resistance that is reversed by the mTOR/phosphatidylinositol 3-kinase inhibitor NVP-BEZ235. Cancer Res. 2008, 68, 9221–9230. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, Y.; Mukohara, T.; Shimada, H.; Saijo, N.; Hirai, M.; Minami, H. Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Ann. Oncol. 2010, 21, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Dupont Jensen, J.; Laenkholm, A.V.; Knoop, A.; Ewertz, M.; Bandaru, R.; Liu, W.; Hackl, W.; Barrett, J.C.; Gardner, H. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin. Cancer Res. 2011, 17, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.; Krishnakumar, S.; Powell, A.A.; Zhang, H.; Mindrinos, M.N.; Telli, M.L.; Davis, R.W.; Jeffrey, S.S. Single cell mutational analysis of PIK3CA in circulating tumor cells and metastases in breast cancer reveals heterogeneity, discordance, and mutation persistence in cultured disseminated tumor cells from bone marrow. BMC Cancer 2014, 14, 456. [Google Scholar] [CrossRef] [PubMed]
- Gasch, C.; Oldopp, T.; Mauermann, O.; Gorges, T.M.; Andreas, A.; Coith, C.; Muller, V.; Fehm, T.; Janni, W.; Pantel, K.; et al. Frequent detection of PIK3CA mutations in single circulating tumor cells of patients suffering from HER2-negative metastatic breast cancer. Mol. Oncol. 2016, 10, 1330–1343. [Google Scholar] [CrossRef] [PubMed]
- Aktas, B.; Muller, V.; Tewes, M.; Zeitz, J.; Kasimir-Bauer, S.; Loehberg, C.R.; Rack, B.; Schneeweiss, A.; Fehm, T. Comparison of estrogen and progesterone receptor status of circulating tumor cells and the primary tumor in metastatic breast cancer patients. Gynecol. Oncol. 2011, 122, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Sieuwerts, A.M.; Mostert, B.; Bolt-de Vries, J.; Peeters, D.; de Jongh, F.E.; Stouthard, J.M.; Dirix, L.Y.; van Dam, P.A.; Van Galen, A.; de Weerd, V.; et al. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin. Cancer Res. 2011, 17, 3600–3618. [Google Scholar] [CrossRef] [PubMed]
- Navin, N.E. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015, 25, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Neves, R.P.; Raba, K.; Schmidt, O.; Honisch, E.; Meier-Stiegen, F.; Behrens, B.; Mohlendick, B.; Fehm, T.; Neubauer, H.; Klein, C.A.; et al. Genomic high-resolution profiling of single CKpos/CD45neg flow-sorting purified circulating tumor cells from patients with metastatic breast cancer. Clin. Chem. 2014, 60, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Auer, M.; Gasch, C.; Pichler, M.; Ulz, P.; Hoffmann, E.M.; Lax, S.; Waldispuehl-Geigl, J.; Mauermann, O.; Lackner, C.; et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 2013, 73, 2965–2975. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Zhuo, M.; Su, Z.; Duan, J.; Gao, Y.; Wang, Z.; Zong, C.; Bai, H.; Chapman, A.R.; Zhao, J.; et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc. Natl. Acad. Sci. USA 2013, 110, 21083–21088. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Adalsteinsson, V.A.; Cibulskis, K.; Choudhury, A.D.; Rosenberg, M.; Cruz-Gordillo, P.; Francis, J.M.; Zhang, C.Z.; Shalek, A.K.; Satija, R.; et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 2014, 32, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Dago, A.E.; Stepansky, A.; Carlsson, A.; Luttgen, M.; Kendall, J.; Baslan, T.; Kolatkar, A.; Wigler, M.; Bethel, K.; Gross, M.E.; et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS ONE 2014, 9, e101777. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.A.; Guttery, D.S.; Hills, A.; Fernandez-Garcia, D.; Page, K.; Rosales, B.M.; Goddard, K.S.; Hastings, R.K.; Luo, J.; Ogle, O.; et al. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high CTC counts. Clin. Cancer Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Fribbens, C.; O’Leary, B.; Kilburn, L.; Hrebien, S.; Garcia-Murillas, I.; Beaney, M.; Cristofanilli, M.; Andre, F.; Loi, S.; Loibl, S.; et al. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J. Clin. Oncol. 2016, 34, 2961–2968. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Mani, S.A.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; Weinberg, R.A. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Mishra, L.; Li, S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015, 6, 10697–10711. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Jiang, J.; Liang, X.H.; Tang, Y.L. Links between cancer stem cells and epithelial-mesenchymal transition. Onco Targets Ther. 2015, 8, 2973–2980. [Google Scholar] [PubMed]
- Kondaveeti, Y.; Guttilla Reed, I.K.; White, B.A. Epithelial-mesenchymal transition induces similar metabolic alterations in two independent breast cancer cell lines. Cancer Lett. 2015, 364, 44–58. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.H.; Yook, J.I.; Kim, H.S.; Kim, N.H. Catabolic metabolism during cancer EMT. Arch. Pharm. Res. 2015, 38, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Metabolic reprogramming: The emerging concept and associated therapeutic strategies. J. Exp. Clin. Cancer Res. 2015, 34, 111. [Google Scholar] [CrossRef] [PubMed]
- Esteban, M.A.; Tran, M.G.; Harten, S.K.; Hill, P.; Castellanos, M.C.; Chandra, A.; Raval, R.; O’Brien, T.S.; Maxwell, P.H. Regulation of E-cadherin expression by VHL and hypoxia-inducible factor. Cancer Res. 2006, 66, 3567–3575. [Google Scholar] [CrossRef] [PubMed]
- Pouyssegur, J.; Dayan, F.; Mazure, N.M. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 2006, 441, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Pennacchietti, S.; Michieli, P.; Galluzzo, M.; Mazzone, M.; Giordano, S.; Comoglio, P.M. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell 2003, 3, 347–361. [Google Scholar] [CrossRef]
- Yang, M.H.; Wu, M.Z.; Chiou, S.H.; Chen, P.M.; Chang, S.Y.; Liu, C.J.; Teng, S.C.; Wu, K.J. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell Biol. 2008, 10, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Gogvadze, V.; Orrenius, S.; Zhivotovsky, B. Mitochondria in cancer cells: What is so special about them? Trends Cell Biol. 2008, 18, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Palorini, R.; Votta, G.; Balestrieri, C.; Monestiroli, A.; Olivieri, S.; Vento, R.; Chiaradonna, F. Energy metabolism characterization of a novel cancer stem cell-like line 3AB-OS. J. Cell. Biochem. 2014, 115, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.A.; Wang, C.Y.; Hsieh, Y.T.; Chen, Y.J.; Wei, Y.H. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle 2015, 14, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Haq, R.; Shoag, J.; Andreu-Perez, P.; Yokoyama, S.; Edelman, H.; Rowe, G.C.; Frederick, D.T.; Hurley, A.D.; Nellore, A.; Kung, A.L.; et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell 2013, 23, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wei, D.; Gao, W.; Xu, Y.; Hu, Z.; Ma, Z.; Gao, C.; Zhu, X.; Li, Q. TPO-Induced Metabolic Reprogramming Drives Liver Metastasis of Colorectal Cancer CD110+ Tumor-Initiating Cells. Cell Stem Cell 2015, 17, 47–59. [Google Scholar] [CrossRef] [PubMed]
| Method for CTC Detection | Subset of CTC Identified | Clinical Relevance | References |
|---|---|---|---|
| CellSearch assay (Veridex) | EpCAM+/CK8+/CD45− | A number of CTC ≥5 cells/7.5 mL at baseline and at the first follow-up represented an independent negative prognostic factor for OS and PFS. | [8,28,29,33,34] |
| EpCAM+/CK+/CD45− | An elevated CTC number before the second cycle of chemotherapy was an early predictive marker of poor PFS and OS. | [35] | |
| EpCAM+/CK+/CD45−/M30−/+ | The presence of M30-negative CTC was associated with a decreased chance of survival in metastatic patients. Both a decrease in the total CTC number and an increase in the fraction of apoptotic CTC (M30-positive) represented a predictive marker. | [36] | |
| EpCAM+/CK+/CD45−/HER2± | Evidence that HER2-negative primary tumors could develop HER2-positive CTC during disease progression. The HER2 status of CTC could be a prognostic factor in MBC patients. | [30,37,38,39,40] | |
| Adna Test Breast Cancer | MUCIN-1-EpCAM+/HER2± | HER2-positive CTC could be detected in HER2-negative primary tumors. | [41] |
| MUCIN-1-EpCAM+/Twist1±/Akt2±/Pl3Kα±/ALDH1± | CTC expressing EMT or stem cell-like markers were associated with poor prognosis and drug resistance. | [42] | |
| MUCIN-1-EpCAM+/HER2±/ER±/PgR± | The molecular profiling of CTC could predict the risk of recurrence and drug resistance. | [43] | |
| DEPArray (Silicon Biosystems) | EpCAM+/CK+/CD45− | The mutational analysis of the TP53 status of CTC showed the presence of heterogeneity between CTC and primary tumors. The presence of TP53 mutations, as assessed by next-generation sequencing performed on single-cell sorted CTC, could represent a negative prognostic factor. | [44,45] |
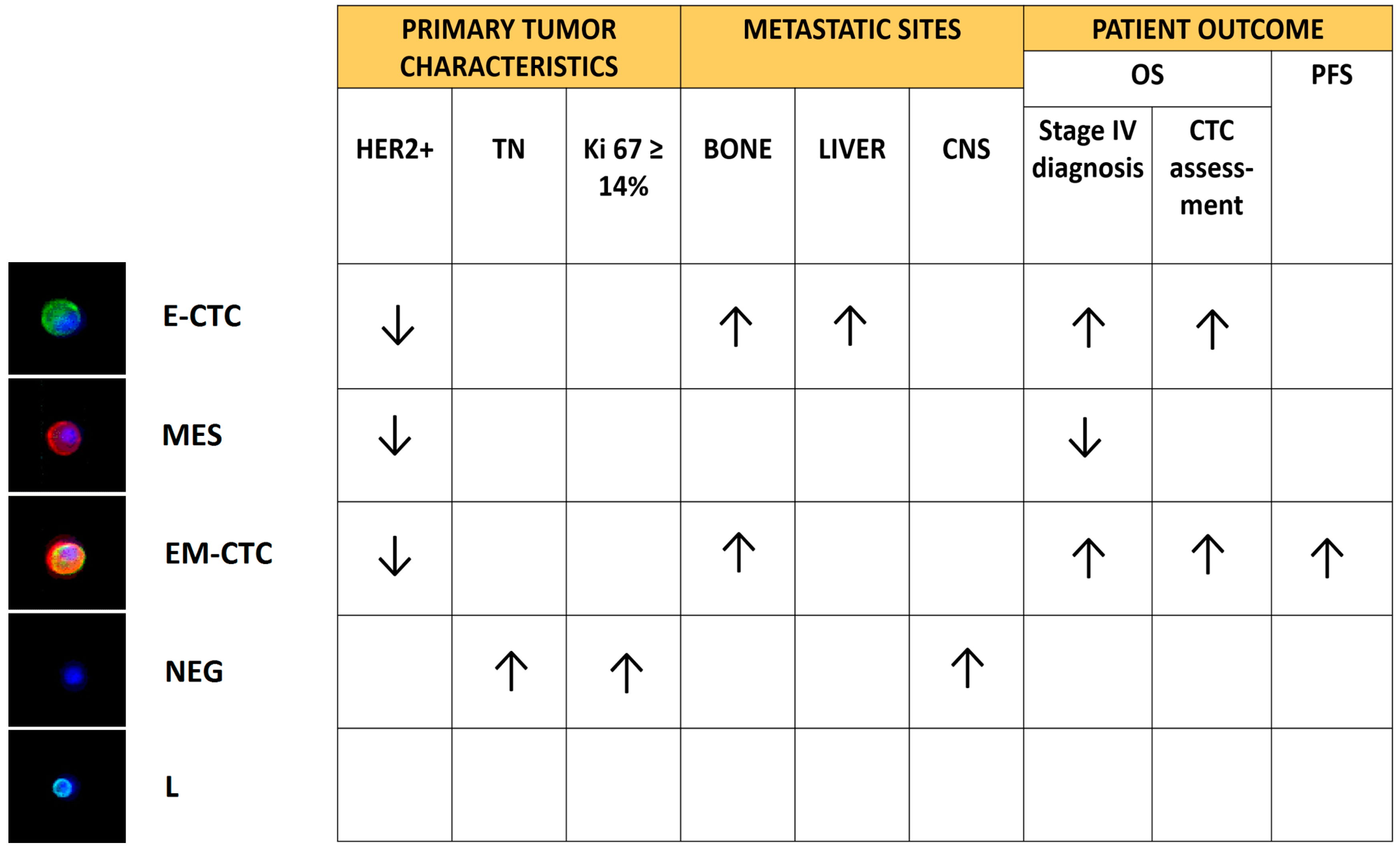
| 4 CD45-negative subsets: Epithelial-CTC: EpCAM-E-cadherin+ EM-CTC: EpCAM-E-cadherin+/CD44-CD146-N-cadherin+ Mesenchymal cells: CD44-CD146-N-cadherin+/EpCAM−/E-cadherin− Negative cells: EpCAM−/E-cadherin−/CD44−/CD146−/N-cadherin− | The presence of CTC in EMT was associated with a poor prognosis. The study highlighted also a correlation between the clinicopathological features of patients and the different subsets of CTC identified. | [46] | |
| EpCAM+/CK+/CD45− | The presence of activating PIK3CA mutations in CTC could predict resistance to anti-HER2 therapies. | [47,48] | |
| Fluorescence in situ hybridization (FISH) | EpCAM+/CK+/CD45−/HER2 amplification± | Evidence that HER2-negative primary tumors developed HER2-positive CTC during disease progression, opening the way to targeted therapies. | [49] |
| Dual-colorimetric RNA-in situ hybridization | E-CTC: CK5-CK7-CK8-CK18-CK9-EpCAM-E-cadherin+ M-CTC: FN1-N-cadherin-SERPINE1-PAI1+ | The mesenchymal immunophenotype was associated with disease progression. Furthermore, CTC from patients with lobular breast cancers were predominantly epithelial-like, whereas those from the triple negative and HER2-positive subtypes were predominantly mesenchymal-like. | [50] |
| Fluorescence activated cell sorting (FACS) | EpCAM−/HER2+/EGFR+/Heparanase+/Notch1+ | Identification, on CTC, of a signature suggestive of metastatic competency to the brain. | [7] |
| EPISPOT (Epithelial ImmunoSPOT) assay | CK19+/MUCIN-1+ | CTC releasing CK19 (CK19-RC) were correlated to an unfavorable clinical outcome. | [51] |
| ISET (isolation by size of epithelial tumor cells) | Size/CK7+ | Evidence that EpCAM-negative CTC could escape from the CellSearch analysis. | [52] |
| RT-qPCR | CK19 mRNA | CK19 mRNA-positive cells could be detected in both early-stage and metastatic breast cancer patients, suggesting the use of RT-qPCR for the continuous monitoring and quantification of circulating epithelial cells. | [53] |
| EM-CTC: EpCAM+/CD45−/TWIST1+/SNAIL1+/ZEB1+ Cancer stem cell-like cells: EpCAM+/CD45−/ALDH+/CD133+ | EM-CTC and cancer stem cell-like cells had a prognostic value in HER2-positive MBC patients treated with targeted therapies. | [54] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulfoni, M.; Turetta, M.; Del Ben, F.; Di Loreto, C.; Beltrami, A.P.; Cesselli, D. Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack. Int. J. Mol. Sci. 2016, 17, 1775. https://doi.org/10.3390/ijms17101775
Bulfoni M, Turetta M, Del Ben F, Di Loreto C, Beltrami AP, Cesselli D. Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack. International Journal of Molecular Sciences. 2016; 17(10):1775. https://doi.org/10.3390/ijms17101775
Chicago/Turabian StyleBulfoni, Michela, Matteo Turetta, Fabio Del Ben, Carla Di Loreto, Antonio Paolo Beltrami, and Daniela Cesselli. 2016. "Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack" International Journal of Molecular Sciences 17, no. 10: 1775. https://doi.org/10.3390/ijms17101775