miR-92a-3p Exerts Various Effects in Glioma and Glioma Stem-Like Cells Specifically Targeting CDH1/β-Catenin and Notch-1/Akt Signaling Pathways
Abstract
:1. Introduction
2. Results
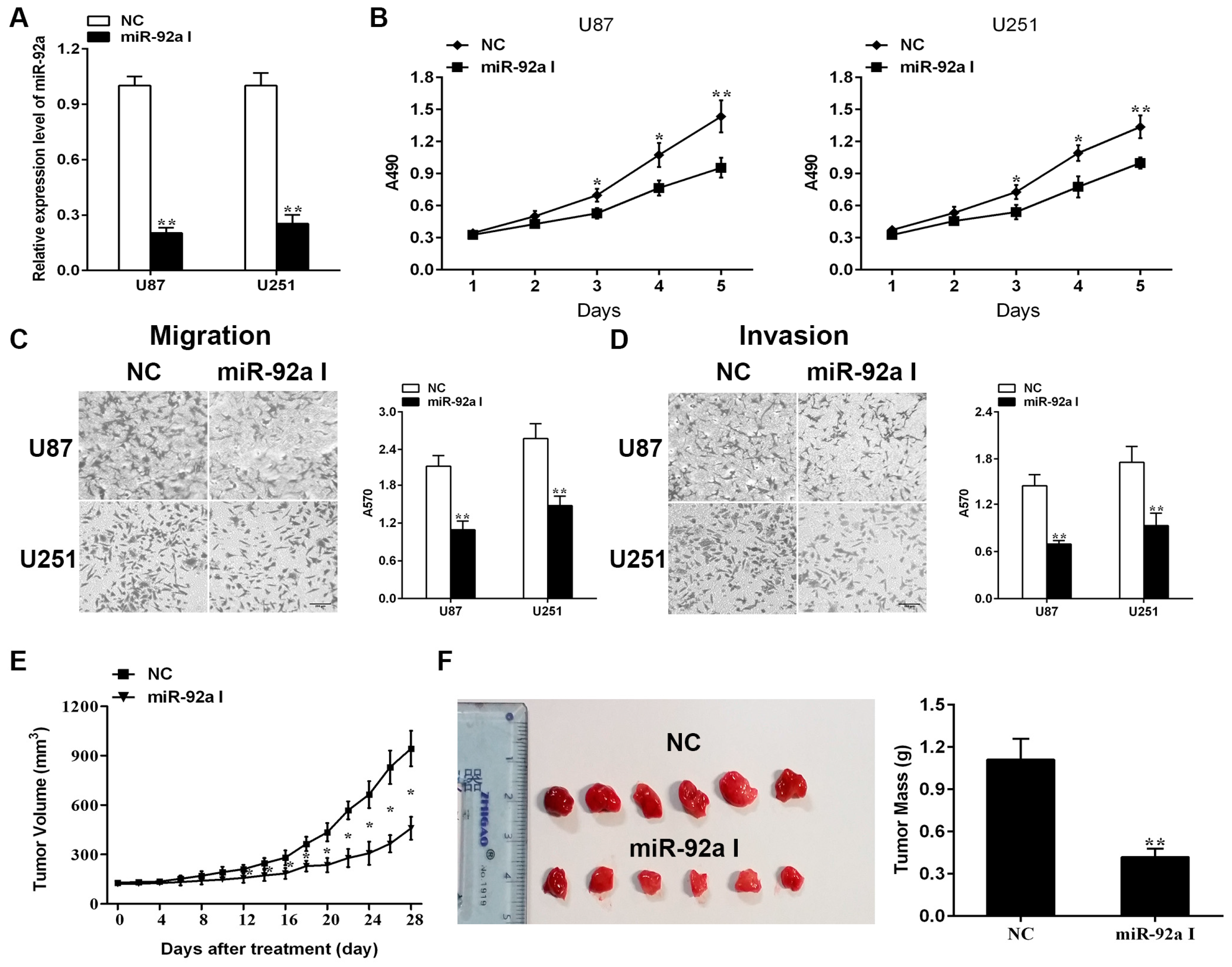
2.1. Depleting Endogenous miR-92a-3p Inhibits the Proliferationand Metastasis of Glioma Cells In Vitro While Preventing Xenograft Growth of Glioma In Vivo
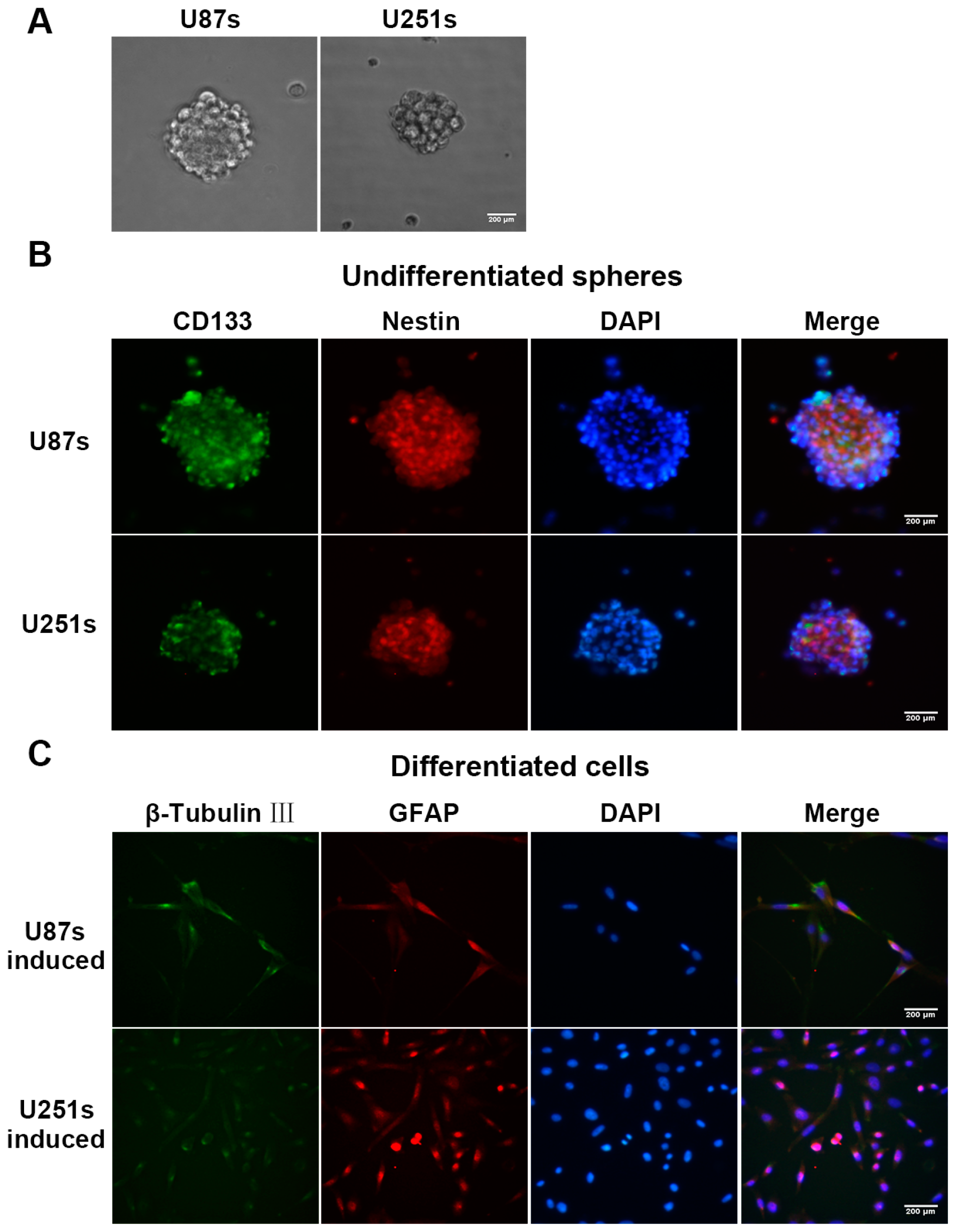
2.2. Isolation and Identification of Glioma Stem-Like Cells (GSCs)
2.3. Over-Expression of miR-92a-3p Suppresses the Self-Renewal Ablity of Glioma Stem-Like Cells
2.4. miR-92a-3p Targets CDH1 in Glioma Cells While Bingding to the 3’UTR of Notch-1 in GSCs
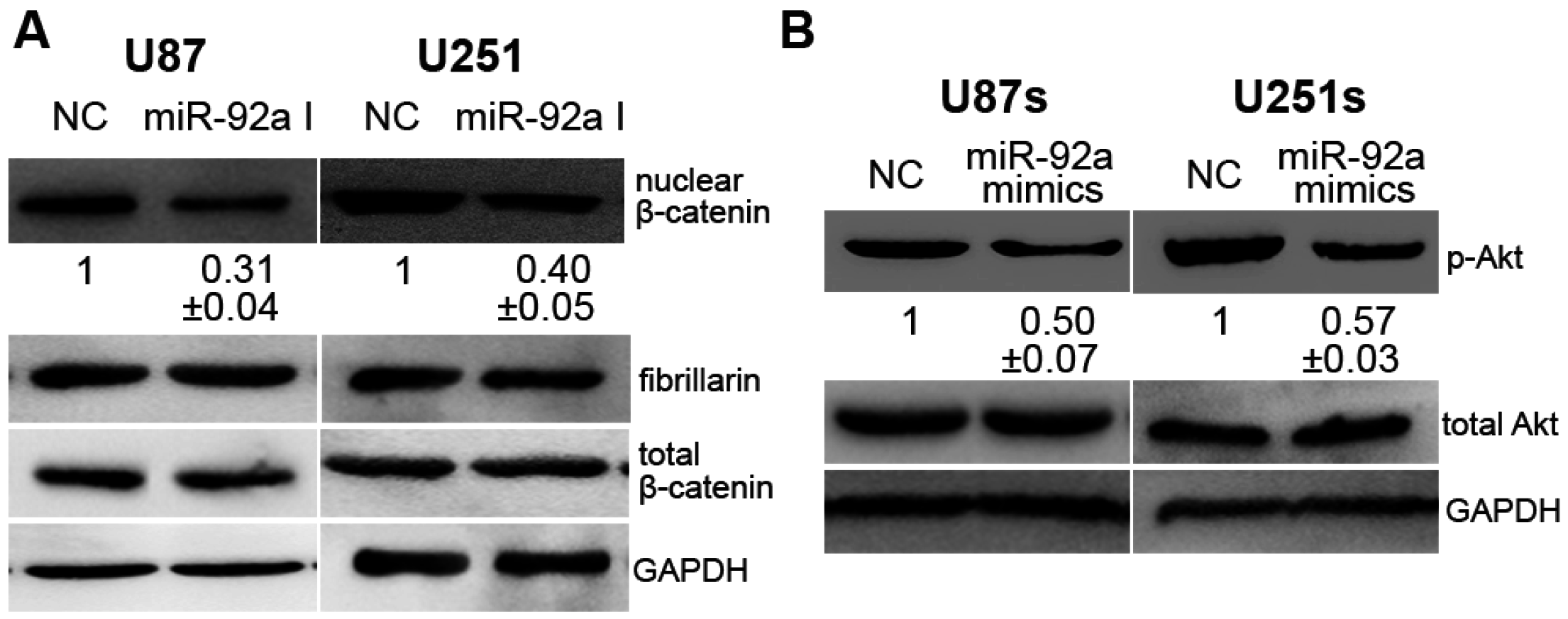
2.5. miR-92a-3p Affected Glioma and GSCs Differently by Acting on CDH1/β-Catenin and Notch-1/Akt Signaling Pathways
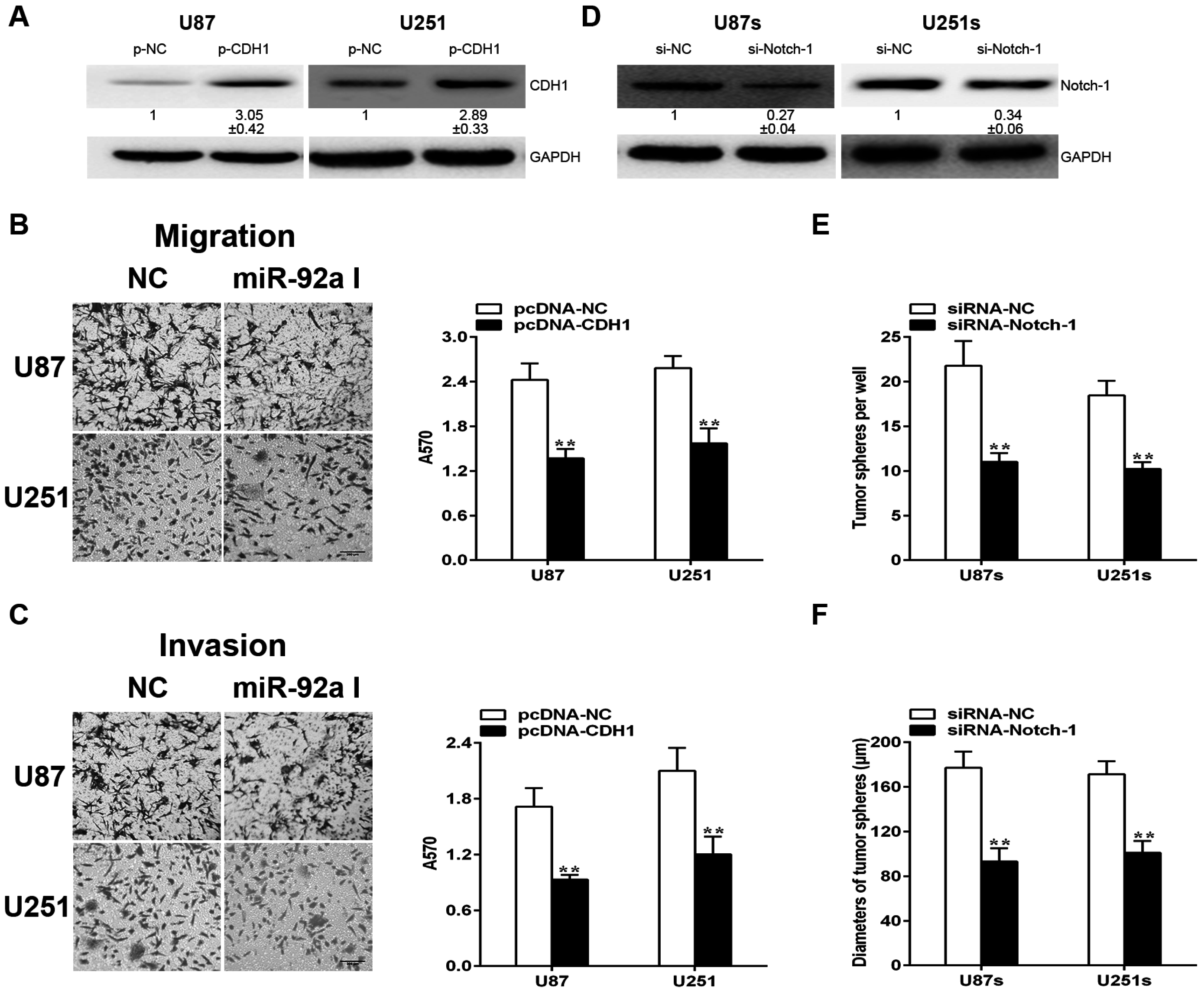
2.6. CDH1and Notch-1 Is Required for miR-92a-3p to Exert Various Effects in Gliomas and GSCs, Respectively
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Differentiation Assay of Tumor Spheres
4.3. Oligonucleotides, Plasmid and Transfection
4.4. RNA Isolation and Quantitative RT-PCR
4.5. Cell Proliferation Assay
4.6. Cell Migration and Invasion Assay
4.7. Tumor Sphere Formation Assay
4.8. Target Prediction
4.9. Reporter Vectors Construction and Luciferase Reporter Assay
4.10. Western Blot Analysis
4.11. Animal Studies
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, T. Understanding high grade glioma: Molecular mechanism, therapy and comprehensive management. Cancer Lett. 2013, 331, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Pollack, I.F. Neuro-oncology: Therapeutic benefits of reirradiation for recurrent brain tumors. Nat. Rev. Neurol. 2010, 6, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, V.; Taylor, M.D. The amazing and deadly glioma race. Cancer Cell 2015, 28, 275–277. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Curtin, J.; Xiong, Y.; Liu, G.; Waschsmann-Hogiu, S.; Farkas, D.L.; Black, K.L.; Yu, J.S. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 2004, 23, 9392–9400. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yuan, X.; Zeng, Z.; Tunici, P.; Ng, H.; Abdulkadir, I.R.; Lu, L.; Irvin, D.; Black, K.L.; Yu, J.S. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 2006. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.; Rich, J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Tuschl, T. Mechanisms of gene silencing by double stranded RNA. Nature 2004, 431, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Bode, A.M.; Cao, Y.; Dong, Z. Regulatory mechanisms and clinical perspectives of miRNA in tumor radiosensitivity. Carcinogenesis 2012, 33, 2220–2227. [Google Scholar] [CrossRef] [PubMed]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Gao, X.; Li, G.; Fu, H.; Cui, D.; Liu, H.; Jin, W.; Zhang, Y. MicroRNA-218 inhibits glioma invasion, migration, proliferation, and cancer stem-like cell self-renewal by targeting the polycomb group gene Bmi1. Cancer Res. 2013, 73, 6046–6055. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Pan, J.S.; Jin, L.X.; Wu, J.; Ren, Y.D.; Chen, P.; Xiao, C.; Han, J. MicroRNA-17–92 inhibits colorectal cancer progression by targeting angiogenesis. Cancer Lett. 2016, 376, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Ma, L.; Zhou, J.; Jiang, M.; Rao, E.; Zhao, Y.; Guo, F. miR-17–92 plays an oncogenic role and conveys chemo-resistance to cisplatin in human prostate cancer cells. Int. J. Oncol. 2016, 48, 1737–1748. [Google Scholar] [CrossRef] [PubMed]
- Ke, T.W.; Wei, P.L.; Yeh, K.T.; Chen, W.T.; Cheng, Y.W. miR-92a promotes cell metastasis of colorectal cancer through PTEN-mediated PI3K/AKT pathway. Ann. Surg. Oncol. 2015, 22, 2649–2655. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Wang, K.; Zhang, A.; Yang, S.; Song, Z.; Wang, W.; Qian, C.; Li, X.; Zhu, Y.; Wang, Y. miR-92a is a critical regulator of the apoptosis pathway in glioblastoma with inverse expression of BCL2L11. Oncol. Rep. 2012, 28, 1771–1777. [Google Scholar] [CrossRef] [PubMed]
- Bentwich, I.; Avniel, A.; Karov, Y.; Aharonov, R.; Gilad, S.; Barad, O.; Barzilai, A.; Einat, P.; Einav, U.; Meiri, E.; et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005, 37, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 12, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Polakis, P. Wnt signaling and cancer. Genes Dev. 2000, 14, 1837–1851. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, J.; Xiong, N.; Li, S.; Chen, Y.; Yang, H.; Wu, C.; Zeng, H.; Liu, Y. Notch-1 signaling activates NF-κB in human breast carcinoma MDA-MB-231 cells via PP2A-dependent AKT pathway. Med. Oncol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Bao, Z.; Wang, Y.; Yang, L.; Lu, B.; Yan, K.; Wang, S.; Wei, H.; Zhang, Z.; Cui, G. Karyopherin β1 regulates proliferation of human glioma cells via Wnt/β-catenin pathway. Biochem. Biophys. Res. Commun. 2016, 478, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Bhuvanalakshmi, G.; Arfuso, F.; Millward, M.; Dharmarajan, A.; Warrier, S. Secreted frizzled-related protein 4 inhibits glioma stem-like cells by reversing epithelial to mesenchymal transition, inducing apoptosis and decreasing cancer stem cell properties. PLoS ONE 2015, 10, e0127517. [Google Scholar]
- Liu, M.; Wang, J.; Huang, B.; Chen, A.; Li, X. Oleuropein inhibits the proliferation and invasion of glioma cells via suppression of the AKT signaling pathway. Oncol. Rep. 2016, 36, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Clark, P.A.; Bhattacharya, S.; Elmayan, A.; Darjatmoko, S.R.; Thuro, B.A.; Yan, M.B.; van Ginkel, P.R.; Polans, A.S.; Kuo, J.S. Resveratrol targeting of AKT and p53 in glioblastoma and glioblastoma stem-like cells to suppress growth and infiltration. J. Neurosurg. 2016, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, A.; Ohno, S.; Wu, W.; Borjigin, N.; Fujita, K.; Aoki, T.; Ueda, S.; Takanashi, M.; Kuroda, M. miR-92 is a key oncogenic component of the miR-17–92 cluster in colon cancer. Cancer Sci. 2011, 102, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Shigoka, M.; Tsuchida, A.; Matsudo, T.; Nagakawa, Y.; Saito, H.; Suzuki, Y.; Aoki, T.; Murakami, Y.; Toyoda, H.; Kumada, T.; et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol. Int. 2010, 60, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.L.; Zhao, X.H.; Wang, J.W.; Li, B.Z.; Wang, Z.; Sun, J.; Tan, F.W.; Ding, D.P.; Xu, X.H.; Zhou, F.; et al. microRNA-92a promotes lymph node metastasis of human esophageal squamous cell carcinoma via E-cadherin. J. Biol. Chem. 2011, 286, 10725–10734. [Google Scholar] [CrossRef] [PubMed]
- Fogel, G.B.; Kai, Z.S.; Zargar, S.; Hinton, A.; Jones, G.A.; Wong, A.S.; Ficici, S.G.; Lopez, A.D.; King, C.C. MicroRNA dynamics during human embryonic stem cell differentiation to pancreatic endoderm. Gene 2015, 574, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Shaham, L.; Binder, V.; Gefen, N.; Borkhardt, A.; Izraeli, S. miR-125 in normal and malignant hematopoiesis. Leukemia 2012, 26, 2011–2018. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.M.; Lin, K.Y.; Chen, Y.Q. Diverse functions of miR-125 family in different cell contexts. J. Hematol. Oncol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Aldaz, B.; Sagardoy, A.; Nogueira, L.; Guruceaga, E.; Grande, L.; Huse, J.T.; Aznar, M.A.; Díez-Valle, R.; Tejada-Solís, S.; Alonso, M.M.; et al. Involvement of miRNAs in the differentiation of human glioblastoma multiforme stem-like cells. PLoS ONE 2013, 8, e77098. [Google Scholar] [CrossRef] [PubMed]
- Canel, M.; Serrels, A.; Frame, M.C.; Brunton, V.G. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J. Cell Sci. 2013, 126, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Onder, T.T.; Gupta, P.B.; Mani, S.A.; Yang, J.; Lander, E.S.; Weinberg, R.A. Loss of Ecadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008, 68, 3645–3654. [Google Scholar] [CrossRef] [PubMed]
- Nelson, W.J.; Nusse, R. Convergence of Wnt, β-catenin, and cadherin pathways. Science 2004, 303, 1483–1487. [Google Scholar] [CrossRef] [PubMed]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial andmesenchymal states: Acquisition of malignant and stem celltraits. Nat. Rev. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Hashimoto, Y.; Kan, T.; Kawamura, J.; Okumura, T.; Soma, T.; Kondo, K.; Teratani, N.; Watanabe, G.; Ino, Y.; et al. Prognostic significance of dysadherin expression in esophageal squamous cell carcinoma. Oncology 2004, 67, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Giles, R.H.; van Es, J.H.; Clevers, H. Caught up in a Wnt storm: Wnt signalingin cancer. Biochim. Biophys. Acta 2003, 1653, 1–24. [Google Scholar] [CrossRef]
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.Y.; Yi, Y.H.; Chang, K.P.; Chang, Y.S.; Chen, S.J.; Chen, H.C. The Epstein-Barr virus-encoded microRNA miR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma. PLoS Pathog. 2014, 10, 852. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Fortini, M.E. Notch signaling: The core pathway and its posttranslational regulation. Dev. Cell 2009, 16, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Bolos, V.; Grego-Bessa, J.; de la Pompa, J.L. Notch signaling in development and cancer. Endocr. Rev. 2007, 28, 339–363. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.L.; Tian, Y.; Lu, C.; Guo, H.; Zhao, X.W.; Guo, Y.W.; Wang, L.Q.; Du, Q.L.; Liu, C.P. Effects of Notch-1 down-regulation on malignant behaviors of breast cancer stem cells. J. Huazhong Univ. Sci. Technol. Med. Sci. 2014, 34, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Qiu, M.; Zhang, Z.; Kang, C.; Jiang, R.; Jia, Z.; Wang, G.; Jiang, H.; Pu, P. The oncogenic roles of Notch1 in astrocytic gliomas in vitro and in vivo. J. Neuro. Oncol. 2010, 97, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhou, Q.; Liu, C.; Tao, M.L. Drug screening study using glioma stem-like cells. Mol. Med. Rep. 2012, 6, 1117–1120. [Google Scholar] [CrossRef] [PubMed]
- Gottardi, C.J.; Wong, E.; Gumbiner, B.M. E-cadherin suppresses cellular transformation by inhibiting β-catenin signaling in an adhesion-independent manner. J. Cell Biol. 2001, 153, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- TargetScan. Available online: http://www.targetscan.org/vert_61/ (accessed on 15 December 2015).
- Microrna.org. Available online: http://www.microrna.org/microrna/home.do (accessed on 15 December 2015).
- National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals, 8th ed.National Academies Press: Washington, DC, USA, 2011; pp. 11–104.


| Oligonucleotides | Sequence 5′ to 3′ |
|---|---|
| miR-92a-3p inhibitor | ACAGGCCGGGACAAGUGCAAUA |
| negative control inhibitor | UCUACUCUUUCUAGGAGGUUGUGA |
| miR-92a-3p mimics | UAUUGCACUUGUCCCGGCCUGU |
| negative control mimics | UCACAACCUCCUAGAAAGAGUAGA |
| Notch-1 siRNA | UGGCGGGAAGUGUGAAGCG |
| negative control siRNA | UUCUCCGAACGUGUCACGU |
| Gene | Primer | Sequence 5′ to 3′ |
|---|---|---|
| miR-92-3p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAGGC |
| Forward | GGGGCAGTTATTGCACTTGTC | |
| Reverse | CCAGTGCAGGGTCCGAGGTA | |
| U6 | RT | AAAAATATGGAACGCTCACGAATTTG |
| Forward | GTGCTCGCTTCGGCAGCACATATAC | |
| Reverse | AAAAATATGGAACGCTCACGAATTTG | |
| CDH1 | Forward | AAAGGCCCATTTCCTAAAAACCT |
| Reverse | TGCGTTCTCTATCCAGAGGCT | |
| Notch-1 | Forward | GAGGCGTGGCAGACTATGC |
| Reverse | CTTGTACTCCGTCAGCGTGA | |
| β-Actin | Forward | AAAGACCTGTACGCCAACAC |
| Reverse | GTCATACTCCTGCTTGCTGAT |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, H.; Zhang, Y.; Liu, N.; Zhao, S.; Kong, Y.; Yuan, L. miR-92a-3p Exerts Various Effects in Glioma and Glioma Stem-Like Cells Specifically Targeting CDH1/β-Catenin and Notch-1/Akt Signaling Pathways. Int. J. Mol. Sci. 2016, 17, 1799. https://doi.org/10.3390/ijms17111799
Song H, Zhang Y, Liu N, Zhao S, Kong Y, Yuan L. miR-92a-3p Exerts Various Effects in Glioma and Glioma Stem-Like Cells Specifically Targeting CDH1/β-Catenin and Notch-1/Akt Signaling Pathways. International Journal of Molecular Sciences. 2016; 17(11):1799. https://doi.org/10.3390/ijms17111799
Chicago/Turabian StyleSong, Hang, Yao Zhang, Na Liu, Sheng Zhao, Yan Kong, and Liudi Yuan. 2016. "miR-92a-3p Exerts Various Effects in Glioma and Glioma Stem-Like Cells Specifically Targeting CDH1/β-Catenin and Notch-1/Akt Signaling Pathways" International Journal of Molecular Sciences 17, no. 11: 1799. https://doi.org/10.3390/ijms17111799





