Comparative Transcriptional Analysis of Loquat Fruit Identifies Major Signal Networks Involved in Fruit Development and Ripening Process
Abstract
:1. Introduction
2. Results
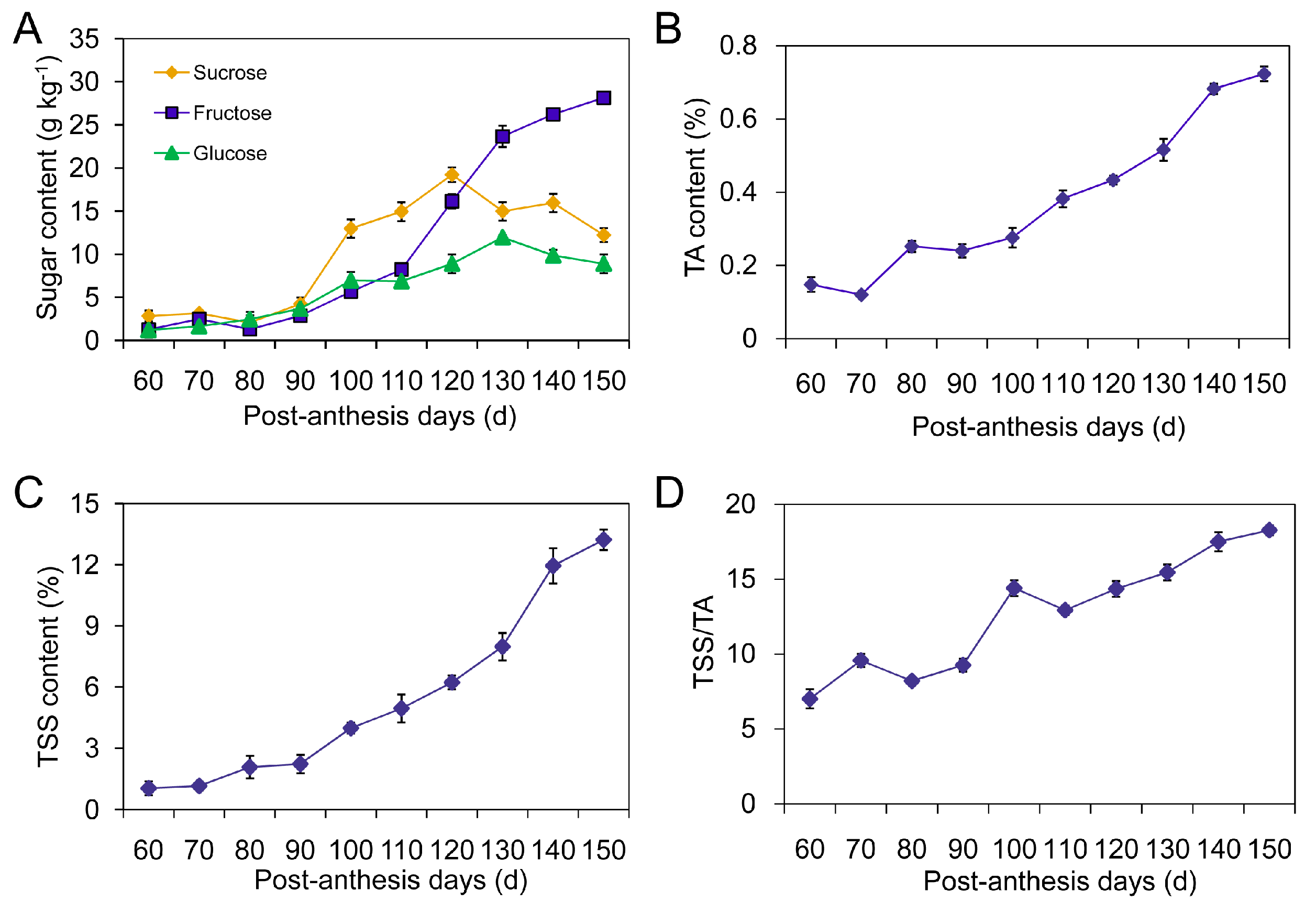
2.1. Changes in Sugar Components, Total Soluble Solids (TSS) and Titratable Acidity (TA)
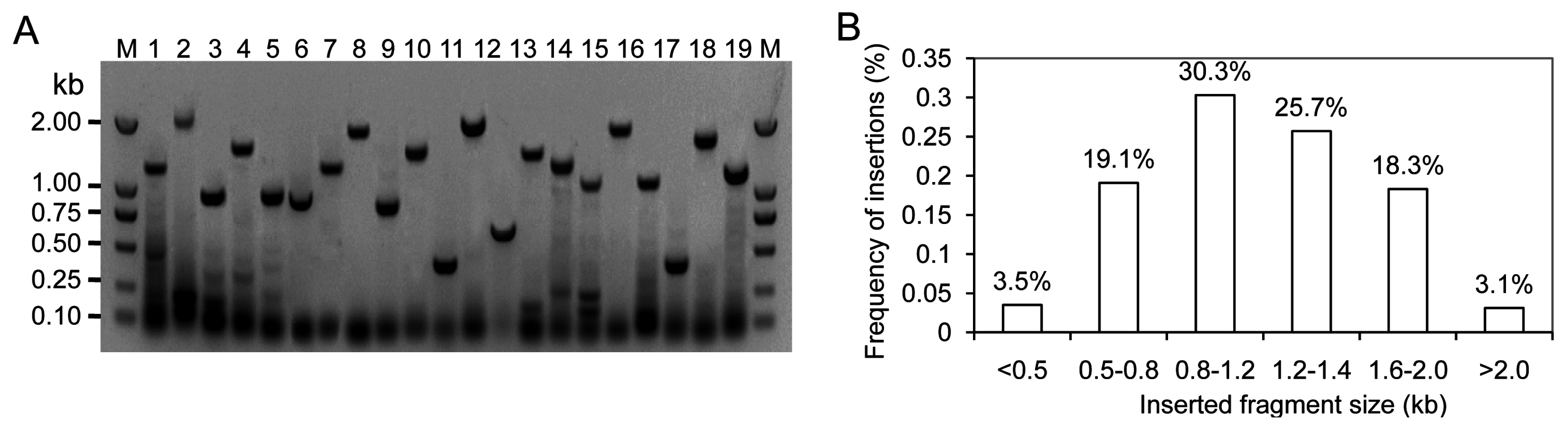
2.2. Generation of a Full-Length Enriched and Normalized cDNA Library
2.3. Transcriptomic Analysis of Loquat Fruit
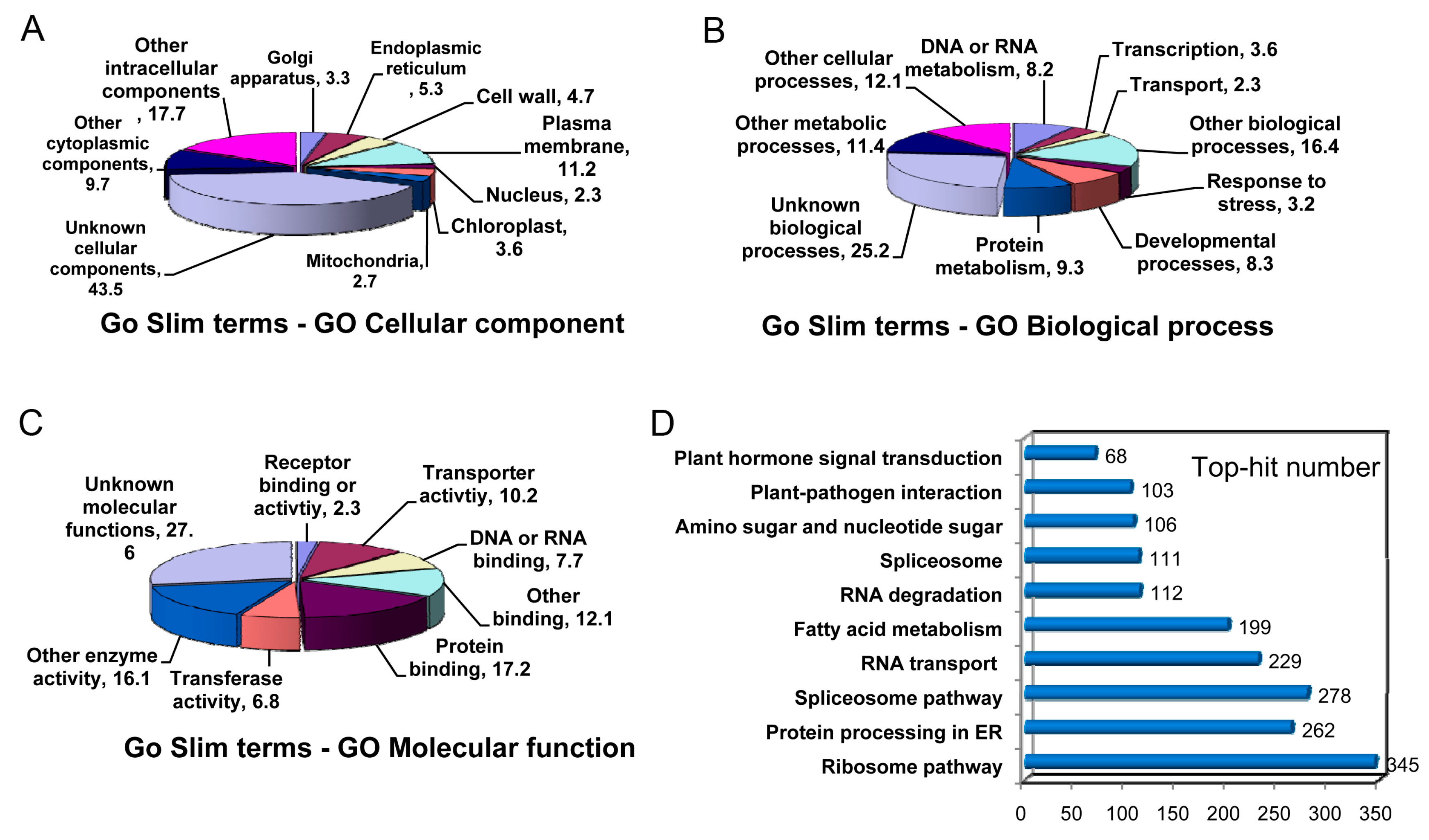
2.4. Sequence Annotation
2.5. Differentially Expressed Genes (DEGs) Involved in Fruit Development and Ripening
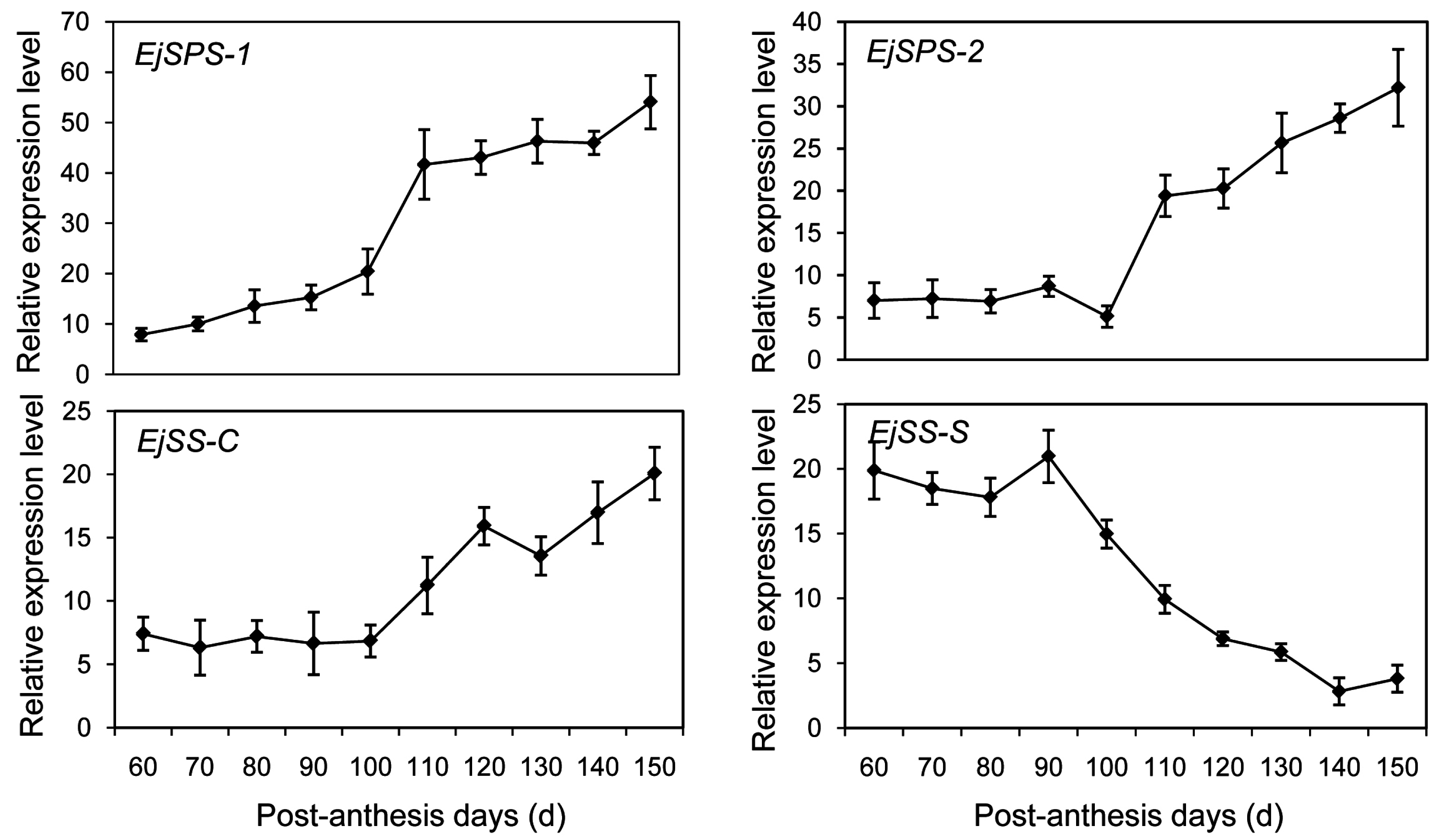
2.6. DEGs Enriched in the Sugar-Metabolism Signalling Pathway
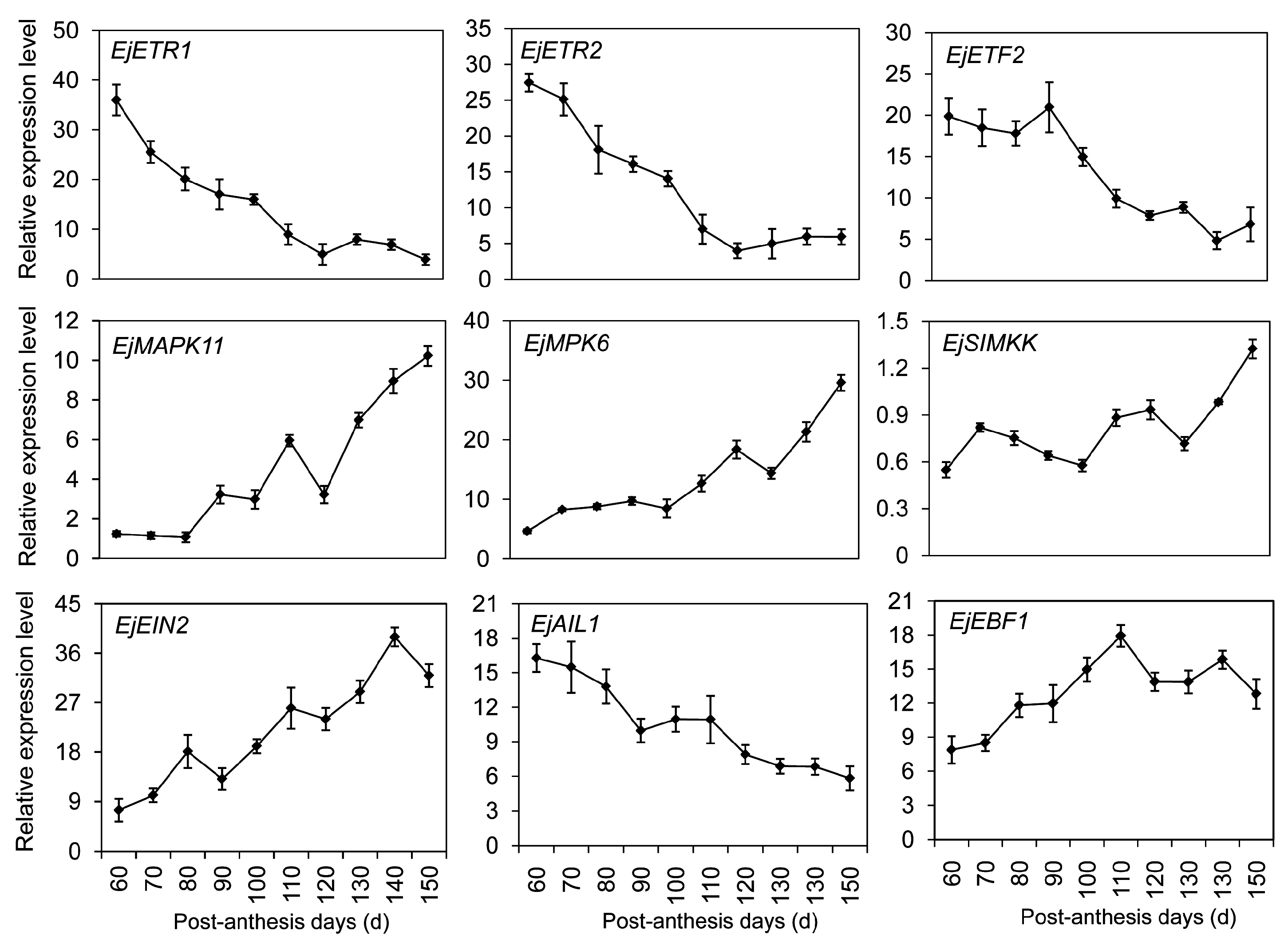
2.7. DEGs Significantly Enriched in the Hormone-Mediated Signaling Pathways
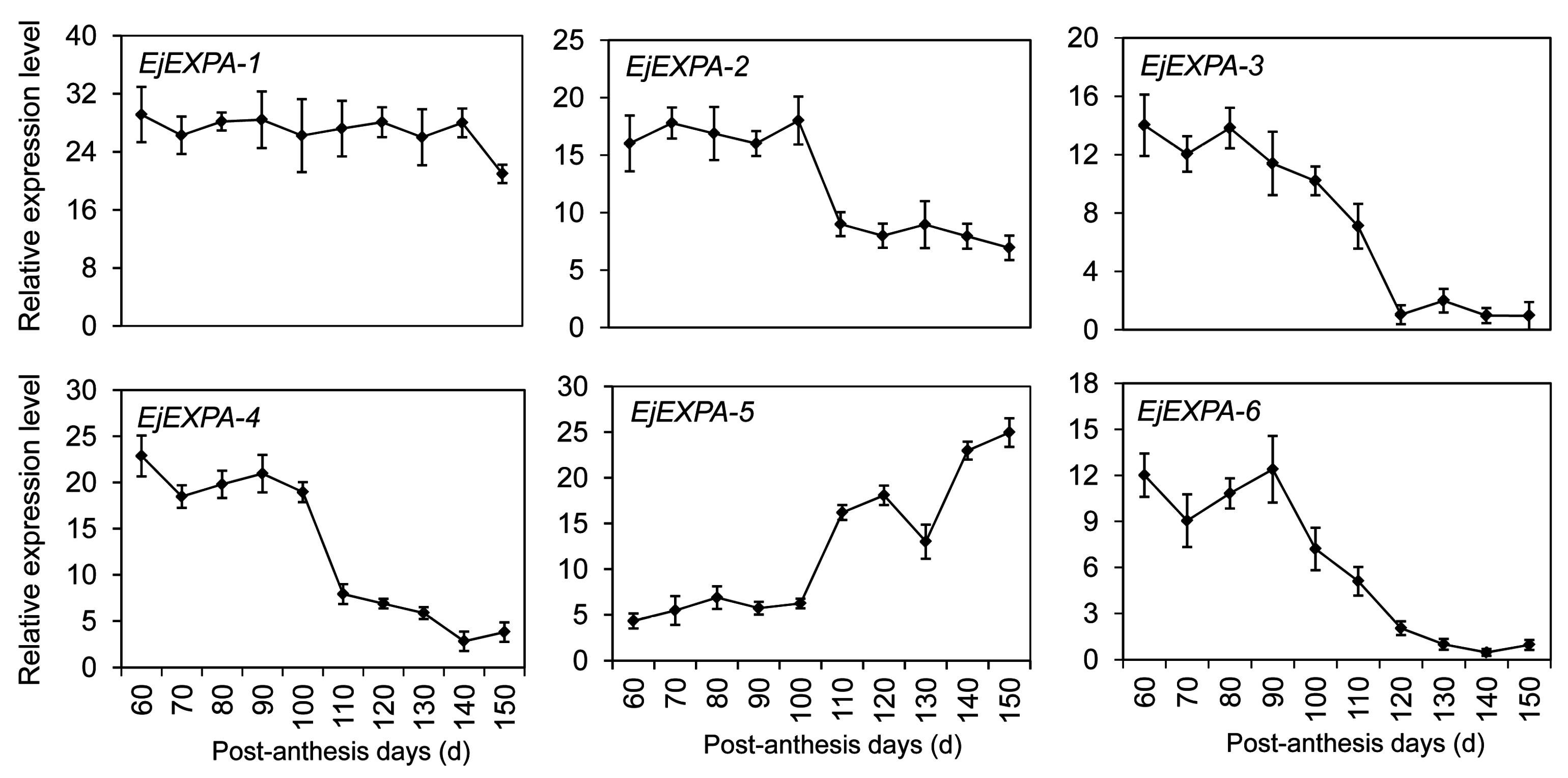
2.8. Expansin Genes Related to Fruit Development
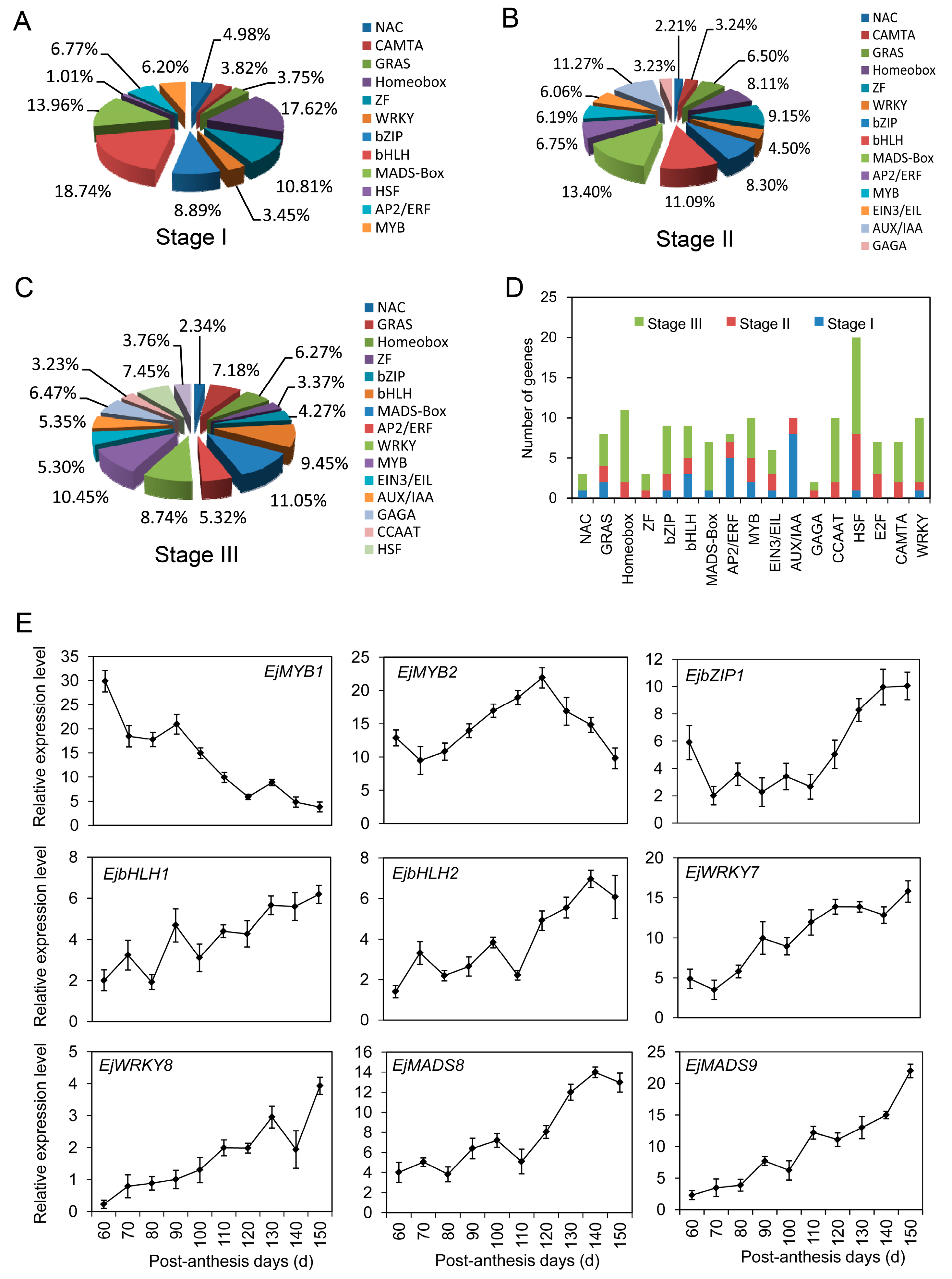
2.9. Transcription Factors Involved in Fruit Development and Ripening
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Sugar Content
4.3. Measurement of TSS and TA
4.4. RNA Isolation and cDNA Synthesis
4.5. First-Strand cDNA Normalization and Single-Strand DNA Amplification
4.6. cDNA Library Construction and Illumina Sequencing
4.7. Sequence Processing and Annotation
4.8. Gene Expression Analysis by qRT-PCR
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, F.X.; Liu, X.H.; Chen, L.S. Developmental changes in pulp organic acid concentration and activities of acid-metabolising enzymes during the fruit development of two loquat (Eriobotrya japonica Lindl.) cultivars differing in fruit acidity. Food Chem. 2009, 114, 657–664. [Google Scholar] [CrossRef]
- Shaw, P.E.; Wilson, C.W. Determination of organic acids and sugars in loquat (Eriobotrya japonica Lindl.) by high-pressure liquid chromatography. J. Sci. Food Agric. 1981, 32, 1242–1246. [Google Scholar] [CrossRef] [PubMed]
- Lanahan, M.B.; Yen, H.C.; Giovannoni, J.J.; Klee, H.J. The never ripe mutation blocks ethylene perception in tomato. Plant Cell 1994, 6, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Drake, R.; Schuch, W.; Giovannoni, J.J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Sun, Y.; Zhang, M.; Wang, L.; Ren, J.; Cui, M.; Wang, Y.; Ji, K.; Li, P.; Li, Q.; et al. Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol. 2012, 158, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Joung, J.G.; McQuinn, R.; Chung, M.Y.; Fei, Z.; Tieman, D.; Klee, H.; Giovannoni, J. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 2012, 70, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Goulas, V.; Minas, I.S.; Kourdoulas, P.M.; Vicente, A.R.; Manganaris, G.A. Phytochemical content, antioxidants and cell wall metabolism of two loquat (Eriobotrya japonica) cultivars under different storage regimes. Food Chem. 2014, 155, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Amorós, A.; Zapata, P.; Pretel, M.T.; Botella, M.A.; Serrano, M. Physico-chemical and physiological changes during fruit development and ripening of five loquat (Eriobotrya japonica Lindl.) cultivars. Food Sci. Technol. Int. 2003, 9, 43–51. [Google Scholar] [CrossRef]
- Cao, S.; Zheng, Y.; Wang, K.; Jin, P.; Rui, H. Methyl jasmonate reduces chilling injury and enhances antioxidant enzyme activity in postharvest loquat fruit. Food Chem. 2009, 115, 1458–1463. [Google Scholar] [CrossRef]
- Ding, Z.; Tian, S.; Wang, Y.; Li, B.; Chan, Z.; Han, J.; Xu, Y. Physiological response of loquat fruit to different storage conditions and its storability. Postharvest Biol. Technol. 2006, 41, 143–150. [Google Scholar] [CrossRef]
- Pareek, S.; Benkeblia, N.; Janick, J.; Cao, S.; Yahia, E.M. Postharvest physiology and technology of loquat (Eriobotrya japonica Lindl.) fruit. J. Sci. Food Agric. 2014, 94, 1495–1504. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Calvo, J.; Gisbert, A.D.; Alamar, M.C.; Hernandorena, R.; Romero, C.; Llácer, G.; Badenes, M.L. Study of a germplasm collection of loquat (Eriobotrya japonica Lindl.) by multivariate analysis. Genet. Resour. Crop Evol. 2008, 55, 695–703. [Google Scholar] [CrossRef]
- Jiang, T.M.; Wang, P.; Yin, X.R.; Zhang, B.; Xu, C.J.; Li, X.; Chen, K.S. Ethylene biosynthesis and expression of related genes in loquat fruit at different developmental and ripening stages. Sci. Hortic. 2011, 130, 452–458. [Google Scholar] [CrossRef]
- Xu, Q.; Yin, X.R.; Zeng, J.K.; Ge, H.; Song, M.; Xu, C.J.; Li, X.; Ferguson, I.B.; Chen, K.S. Activator-and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 2014, 65, 4349–4359. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Sun, C.; Wang, P.; Shan, L.; Cai, C.; Zhang, B.; Zhang, W.; Li, X.; Ferguson, I.; Chen, K. Expression of expansin genes during postharvest lignifications and softening of ‘Luoyangqing’ and ‘Baisha’ loquat fruit under different storage conditions. Postharvest Biol. Technol. 2008, 49, 46–53. [Google Scholar] [CrossRef]
- Zeng, J.K.; Li, X.; Xu, Q.; Chen, J.Y.; Yin, X.R.; Ferguson, I.B.; Chen, K.S. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 2015, 13, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.Y.; Machleder, E.M.; Chenchik, A.; Li, R.; Siebert, P.D. Reverse transcriptase template switching: A SMART™ approach for full-length cDNA library construction. Biotechniques 2001, 30, 892–897. [Google Scholar] [PubMed]
- Rowland, L.J.; Alkharouf, N.; Darwish, O.; Ogden, E.L.; Polashock, J.J.; Bassil, N.V.; Main, D. Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol. 2012, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Chen, M.; Xu, C.J.; Bai, L.; Yin, X.R.; Li, X.; Allan, A.C.; Ferguson, I.B.; Chen, K.S. Transcriptomic analysis of Chinese bayberry (Myrica rubra) fruit development and ripening using RNA-Seq. BMC Genom. 2012, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Xu, Q.; Da, X.; Guo, F.; Ding, Y.; Deng, X. Transcriptome changes during fruit development and ripening of sweet orange (Citrus sinensis). BMC Genom. 2012, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Ibeas, D.; Blanca, J.; Donaire, L.; Saladie, M.; Mascarell-Creus, A.; Cano-Delgado, A.; Garcia-Mas, J.; Llave, C.; Aranda, M.A. Analysis of the melon (Cucumis melo) small RNAome by high-throughput pyrosequencing. BMC Genom. 2011, 12, 393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, A.C.; Boldingh, H.L.; McAtee, P.A.; Gunaseelan, K.; Luo, Z.; Atkinson, R.G.; David, K.M.; Burdon, J.N.; Schaffer, R.J. Fruit development of the diploid kiwifruit, Actinidia chinensis ‘Hort16A’. BMC Plant Biol. 2011, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- BLAST. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome (accessed on 18 May 2016).
- Cherian, S.; Figueroa, C.R.; Nair, H. ‘Movers and shakers’ in the regulation of fruit ripening: A cross-dissection of climacteric versus non-climacteric fruit. J. Exp. Bot. 2014, 65, 4705–4722. [Google Scholar] [CrossRef] [PubMed]
- Malundo, T.M.M.; Shewfelt, R.L.; Ware, G.O.; Baldwin, E.A. Sugars and acids influence flavor properties of mango (Mangifera indica). J. Am. Soc. Hortic. Sci. 2001, 126, 115–121. [Google Scholar]
- Saradhuldhat, P.; Paull, R.E. Pineapple organic acid metabolism and accumulation during fruit development. Sci. Hortic. 2007, 112, 297–303. [Google Scholar] [CrossRef]
- Song, H.W.; Yuan, W.M.; Jin, P.; Wang, W.; Wang, X.F.; Yang, L.M.; Zhang, Y.F. Effects of chitosan/nano-silica on postharvest quality and antioxidant capacity of loquat fruit during cold storage. Postharvest Biol. Technol. 2016, 119, 41–48. [Google Scholar] [CrossRef]
- Ding, C.K.; Chachin, K.; Hamauzu, Y.; Ueda, Y.; Imahori, Y. Effects of storage temperatures on physiology and quality of loquat fruit. Postharvest Biol. Technol. 1998, 14, 309–315. [Google Scholar] [CrossRef]
- Hamauzu, Y.; Chachin, K.; Ding, C.K.; Kurooka, H. Differences in surface color, flesh firmness, physiological activity, and some components of loquat fruits picked at various stages of maturity. J. Jpn. Soc. Hortic. Sci. 1997, 65, 859–865. [Google Scholar] [CrossRef]
- Komatsu, A.; Takanokura, Y.; Moriguchi, T.; Omura, M.; Akihama, T. Differential expression of three sucrose-phosphate synthase isoforms during sucrose accumulation in citrus fruits (Citrus unshiu Marc.). Plant Sci. 1999, 140, 169–178. [Google Scholar] [CrossRef]
- Langenkämper, G.; McHale, R.; Gardner, R.C.; MacRae, E. Sucrose-phosphate synthase steady-state mRNA increases in ripening kiwifruit. Plant Mol. Biol. 1998, 36, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Araújo, W.L.; Nunes-nesi, A.; Nikoloski, Z.; Sweetlove, L.J.; Fernie, A.R. Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 2012, 35, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Kleeff, P.J.; Oecking, C.; Li, K.W.; Erban, A.; Kopka, J.; Hincha, D.K.; Boer, A.H. Light modulated activity of root alkaline/neutral invertase involves the interaction with 14-3-3 proteins. Plant J. 2014, 80, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.C.; Zhao, Y. Auxin perception and downstream events. Curr. Opin. Plant Biol. 2016, 33, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Schauer, N.; Usadel, B.; Frasse, P.; Zouine, M.; Hernould, M.; Latche, A.P.; Bouzayen, M. Regulatory features underlying pollination-dependent and-independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 2009, 21, 1428–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrani, J.C.; Ruiz-Rivero, O.; Fos, M.; García-Martínez, J.L. Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J. 2008, 56, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.; Frasse, P.; Olmos, E.; Zegzouti, H.; Li, Z.G.; Latché, A.; Pech, J.C.; Bouzayen, M. Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J. 2002, 32, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Mounet, F.; Moing, A.; Kowalczyk, M.; Rohrmann, J.; Petit, J.; Garcia, V.; Maucourt, M.; Yano, K.; Deborde, C.; Aoki, K.; et al. Down-regulation of a single auxin efflux transport protein in tomato induces precocious fruit development. J. Exp. Bot. 2012, 63, 4901–4917. [Google Scholar] [CrossRef] [PubMed]
- Paponov, I.A.; Teale, W.D.; Trebar, M.; Blilou, I.; Palme, K. The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci. 2005, 10, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Trainotti, L.; Pavanello, A.; Casadoro, G. Different ethylene receptors show an increased expression during the ripening of strawberries: does such an increment imply a role for ethylene in the ripening of these non-climacteric fruits? J. Exp. Bot. 2005, 56, 2037–2046. [Google Scholar] [CrossRef] [PubMed]
- Cara, B.; Giovannoni, J.J. Molecular biology of ethylene during tomato fruit development and maturation. Plant Sci. 2008, 175, 106–113. [Google Scholar] [CrossRef]
- Purvis, A.C.; Barmore, C.R. Involvement of ethylene in chlorophyll degradation in peel of citrus fruits. Plant Physiol. 1981, 68, 854–856. [Google Scholar] [CrossRef] [PubMed]
- El-Kereamy, A.; Chervin, C.; Roustan, J.P.; Cheynier, V.; Souquet, J.M.; Moutounet, M.; Raynal, J.; Ford, C.; Latché, A.; Pech, J.; et al. Exogenous ethylene stimulates the long-term expression of genes related to anthocyanin biosynthesis in grape berries. Physiol. Plant. 2003, 119, 175–182. [Google Scholar] [CrossRef]
- El-Sharkawy, I.; Sherif, S.; Mila, I.; Bouzayen, M.; Jayasankar, S. Molecular characterization of seven genes encoding ethylene-responsive transcriptional factors during plum fruit development and ripening. J. Exp. Bot. 2009, 60, 907–922. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Bradford, K.J. Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol. 2000, 124, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; Chapman, N.; David, K.; Angenent, G.C.; Seymour, G.B.; de Maagd, R.A. Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 2014, 65, 4527–4541. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, M.; Shima, Y.; Nakagawa, H.; Kitagawa, M.; Kimbara, J.; Nakano, T.; Kasumi, T.; Ito, Y. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 2014, 26, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- McAtee, P.; Karim, S.; Schaffer, R.J.; David, K. A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front. Plant Sci. 2013, 4, 79. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Dai, C.; Lee, M.M.; Kwak, J.M.; Nam, K.H. BRI1-Associated Receptor Kinase 1 regulates guard cell ABA signaling mediated by Open Stomata 1 in Arabidopsis. Mol. Plant 2016, 9, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Stanković, B.; Vian, A.; Henry-Vian, C.; Davies, E. Molecular cloning and characterization of a tomato cDNA encoding a systemically wound-inducible bZIP DNA-binding protein. Planta 2000, 212, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Berberich, T.; Sano, H.; Kusano, T. Specific association of transcripts of tbzF and tbz17, tobacco genes encoding basic region leucine zipper-type transcriptional activators, with guard cells of senescing leaves and/or flowers. Plant Physiol. 2001, 127, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Parra, R.; Paredes, M.A.; Sanchez-Calle, I.M.; Gomez-Jimenez, M.C. Comparative transcriptional profiling analysis of olive ripe-fruit pericarp and abscission zone tissues shows expression differences and distinct patterns of transcriptional regulation. BMC Genom. 2013, 14, 866. [Google Scholar] [CrossRef] [PubMed]
- Corbacho, J.; Romojaro, F.; Pech, J.C.; Latché, A.; Gomez-Jimenez, M.C. Transcriptomic events involved in melon mature-fruit abscission comprise the sequential induction of cell-wall degrading genes coupled to a stimulation of endo and exocytosis. PLoS ONE 2013, 8, e58363. [Google Scholar] [CrossRef] [PubMed]
- Gil-Amado, J.A.; Gomez-Jimenez, M.C. Transcriptome analysis of mature-fruit abscission control in olive. Plant Cell Physiol. 2013, 54, 244–269. [Google Scholar] [CrossRef] [PubMed]
- Botton, A.; Eccher, G.; Forcato, C.; Ferrarini, A.; Begheldo, M.; Zermiani, M.; Moscatello, S.; Battistelli, A.; Velasco, R.; Ruperti, B.; et al. Signaling pathways mediating the induction of apple fruitlet abscission. Plant Physiol. 2011, 155, 185–208. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, J.J. Genetic regulation of fruit development and ripening. Plant Cell 2004, 16, S170–S180. [Google Scholar] [CrossRef] [PubMed]
- White, P.J. Recent advances in fruit development and ripening: An overview. J. Exp. Bot. 2002, 53, 1995–2000. [Google Scholar] [CrossRef] [PubMed]
- Zhulidov, P.A.; Bogdanova, E.A.; Shcheglov, A.S.; Vagner, L.L.; Khaspekov, G.L.; Kozhemyako, V.B.; Matz, M.V.; Meleshkevitch, E.; Shagin, D.A. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004, 32, e37. [Google Scholar] [CrossRef] [PubMed]
- Müller-Herbst, S.; Wüstner, S.; Mühlig, A.; Eder, D.; Fuchs, T.M.; Held, C.; Ehrenreich, A.; Scherer, S. Identification of genes essential for anaerobic growth of Listeria monocytogenes. Microbiology 2014, 160, 752–765. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.D.; Wiemann, S. KEGGgraph: A graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics 2009, 25, 1470–1471. [Google Scholar] [CrossRef] [PubMed]


| Assembly Metric | Sequences (No.) | Base Pairs (bp) | Mean Length (bp) |
|---|---|---|---|
| Raw sequencing reads | 51,881,623 | 5,019,322,712 | 97 |
| Clean reads | 51,610,234 | 4,850,271,306 | 94 |
| Contigs (≥100 bp) | 453,421 | 125,597,617 | 277 |
| Singletons (≥150 bp) | 32,608 | 20,575,648 | 631 |
| Clusters (≥150 bp) | 16,230 | 18,047,760 | 1112 |
| Total unigenes (≥150 bp) | 48,838 | 38,623,512 | 790 |
| Unigene | Putative Function | Accession No. | Source Organism | E-Value a |
|---|---|---|---|---|
| Contig 5600 | Polyphenol oxidase | AEY79824 | Triticum aestivum | 5.17 × 10−9 |
| Contig 5601 | Pyrophosphate-fructose 6-phosphate | XP008453023 | Cucumis melo | 3.11 × 10−7 |
| Contig 5602 | Ethylene cinnamyl alcohol dehydrogenase | XP017179614 | Malus domestica | 5.84 × 10−7 |
| Contig 5603 | ADP-glucose pyrophosphorylase subunit | ABK97520 | Sorghum bicolor | 3.52 × 10−5 |
| Contig 5604 | Sucrose phosphate synthase | AAL86360 | Actinidia chinensis | 3.24 × 10−9 |
| Contig 5605 | Heat shock protein 70 | NP567510 | Arabidopsis thaliana | 2.25 × 10−6 |
| Contig 5606 | Ethylene response factor | JF815559 | E. japonica | 0.0 |
| Contig 5607 | Expansin | EU123922 | E. japonica | 0.0 |
| Contig 5608 | TFL1-like protein | BAD10969 | Pyrus communis | 7.13 × 10−5 |
| Contig 5609 | Ethylene-responsive transcription factor | JF815559 | E. japonica | 1.84 × 10−11 |
| Contig 5610 | Ethylene receptor | NP_00128083 | M. domestica | 1.56 × 10−7 |
| Contig 5611 | Phenylalanine ammonia lyase | EF685344 | E. japonica | 0.0 |
| Contig 5612 | ATP synthase subunit | NP568203 | A. thaliana | 1.28 × 10−7 |
| Contig 5613 | Fructokinase | JF414124 | E. japonica | 0.0 |
| Contig 5614 | Hexokinase | JF414121 | E. japonica | 0.0 |
| Contig 5615 | Catalase 1 | XP008348924 | M. domestica | 0.0 |
| Contig 5616 | Ribulose-1,5-bisphosphate carboxylase | XM003616402 | Medicago truncatula | 2.84 × 10−5 |
| Contig 5617 | Sucrose synthase | XM002266984 | Vitis vinifera | 0.0 |
| Contig 5618 | Phosphoglycerate kinase | M008384936 | M. domestica | 2.03 × 10−7 |
| Contig 5619 | Lipid transfer protein 4 precursor | AY793558 | Lens culinaris | 5.79 × 10−7 |
| Contig 5620 | Myb transcription factor | XM008366832 | M. domestica | 1.23 × 10−5 |
| Contig 5621 | UDP-glucosyltransferase | DR993941 | M. domestica | 1.10 × 10−9 |
| Contig 5622 | Ethylene response factor (ERF) | JF412350 | M. domestica | 2.22 × 10−8 |
| Contig 5623 | Senescence-related protein | XM002300415 | Populus trichocarpa | 2.05 × 10−6 |
| Contig 5624 | Poly(A)-binding protein | NM001198255 | A. thaliana | 0.0 |
| Contig 5625 | Metallothionein-like protein | AF009959 | M. domestica | 5.89 × 10−7 |
| Contig 5626 | Translation elongation factor | XM008340889 | M. domestica | 1.85 × 10−8 |
| Contig 5627 | DELLA protein | KC434135 | Triticum aestivum | 1.11 × 10−8 |
| Contig 5628 | Vacuolar Ca2+/H+ exchanger | AB012932 | Vigna radiata | 2.09 × 10−10 |
| Contig 5629 | ATP binding protein | XM002521829 | Ricinus communis | 0.0 |
| Contig 5630 | Phosphoric diester hydrolase | NM101237 | A. thaliana | 1.66 × 10−5 |
| Contig 5631 | Serine/threonine-protein kinase | XM008391178 | M. domestica | 2.42 × 10−5 |
| Contig 5632 | Cytochrome C reductase | X79275 | Solanum tuberosum | 2.03 × 10−7 |
| Contig 5633 | 1-Aminocyclopropane-1-carboxylic acid oxidase | AB003514 | Actinidia deliciosa | 1.24 × 10−8 |
| Contig 5634 | Vacuolar ATP synthase subunit G1 | NM180158 | A. thaliana | 1.34 × 10−7 |
| Contig 5635 | β-1,3-Glucanase | AY548364 | M. domestica | 2.24 × 10−8 |
| Contig 5636 | Alcohol dehydrogenase | L23548 | Zea mays | 2.54 × 10−6 |
| Contig 5637 | 14-3-3 Family protein | NM001247178 | S. lycopersicum | 2.45 × 10−5 |
| Contig 5638 | MADS box genes | AJ000759 | M. domestica | 0.0 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, H.; Zhao, X.; Hu, W.; Wang, X.; Shen, T.; Yang, L. Comparative Transcriptional Analysis of Loquat Fruit Identifies Major Signal Networks Involved in Fruit Development and Ripening Process. Int. J. Mol. Sci. 2016, 17, 1837. https://doi.org/10.3390/ijms17111837
Song H, Zhao X, Hu W, Wang X, Shen T, Yang L. Comparative Transcriptional Analysis of Loquat Fruit Identifies Major Signal Networks Involved in Fruit Development and Ripening Process. International Journal of Molecular Sciences. 2016; 17(11):1837. https://doi.org/10.3390/ijms17111837
Chicago/Turabian StyleSong, Huwei, Xiangxiang Zhao, Weicheng Hu, Xinfeng Wang, Ting Shen, and Liming Yang. 2016. "Comparative Transcriptional Analysis of Loquat Fruit Identifies Major Signal Networks Involved in Fruit Development and Ripening Process" International Journal of Molecular Sciences 17, no. 11: 1837. https://doi.org/10.3390/ijms17111837






