Rapid, Sensitive Detection of Bartonella quintana by Loop-Mediated Isothermal Amplification of the groEL Gene
Abstract
:1. Introduction
2. Results
2.1. Confirmation and Detection of B. quintana Loop-Mediated Isothermal Amplification (LAMP) Reaction Products
2.2. The Optimal Temperature for the B. quintana LAMP Assay
2.3. Specificity of the B. quintana LAMP Assay
2.4. Sensitivity of the B. quintana LAMP Assay
2.5. Evaluation of Practical Application of the B. quintana LAMP Assay
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Bacterial Strains and Culture Conditions
4.3. Genomic DNA Extraction
4.4. Design of LAMP Primers
4.5. LAMP Reaction
4.6. Specificity and Sensitivity of the B. quintana LAMP Assay
4.7. Practical Application of the B. quintana LAMP
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Regier, Y.; O Rourke, F.; Kempf, V.A. Bartonella spp.—A chance to establish one health concepts in veterinary and human medicine. Parasites Vectors 2016, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Minnick, M.F.; Anderson, B.E.; Lima, A.; Battisti, J.M.; Lawyer, P.G.; Birtles, R.J. Oroya fever and verruga peruana: Bartonelloses unique to south america. PLoS Negl. Trop. Dis. 2014, 8, e2919. [Google Scholar] [CrossRef] [PubMed]
- Guy, L.; Nystedt, B.; Toft, C.; Zaremba-Niedzwiedzka, K.; Berglund, E.C.; Granberg, F.; Naslund, K.; Eriksson, A.S.; Andersson, S.G. A gene transfer agent and a dynamic repertoire of secretion systems hold the keys to the explosive radiation of the emerging pathogen Bartonella. PLoS Genet. 2013, 9, e1003393. [Google Scholar] [CrossRef] [PubMed]
- Foucault, C.; Brouqui, P.; Raoult, D. Bartonella quintana characteristics and clinical management. Emerg. Infect. Dis. 2006, 12, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Breitschwerdt, E.B.; Maggi, R.G.; Nicholson, W.L.; Cherry, N.A.; Woods, C.W. Bartonella sp. Bacteremia in patients with neurological and neurocognitive dysfunction. J. Clin. Microbiol. 2008, 46, 2856–2861. [Google Scholar] [CrossRef] [PubMed]
- Kooli, I.; Loussaief, C.; Ben Brahim, H.; Aouem, A.; Toumi, A.; Chakroun, M. Bartonella quintana meningoencephalitis in an immunocompetent: Rare case. Pathol. Biol. (Paris) 2014, 62, 342–344. [Google Scholar] [CrossRef] [PubMed]
- Abromaitis, S.; Nelson, C.S.; Previte, D.; Yoon, K.S.; Clark, J.M.; DeRisi, J.L.; Koehler, J.E. Bartonella quintana deploys host and vector temperature-specific transcriptomes. PLoS ONE 2013, 8, e58773. [Google Scholar] [CrossRef] [PubMed]
- Pitassi, L.H.; de Paiva Diniz, P.P.; Scorpio, D.G.; Drummond, M.R.; Lania, B.G.; Barjas-Castro, M.L.; Gilioli, R.; Colombo, S.; Sowy, S.; Breitschwerdt, E.B.; et al. Bartonella spp. Bacteremia in blood donors from campinas, brazil. PLoS Negl. Trop. Dis. 2015, 9, e0003467. [Google Scholar] [CrossRef] [PubMed]
- Abromaitis, S.; Koehler, J.E. The Bartonella quintana extracytoplasmic function sigma factor rpoe has a role in bacterial adaptation to the arthropod vector environment. J. Bacteriol. 2013, 195, 2662–2674. [Google Scholar] [CrossRef] [PubMed]
- Bouhsira, E.; Ferrandez, Y.; Liu, M.; Franc, M.; Boulouis, H.J.; Biville, F. Ctenocephalides felis an in vitro potential vector for five Bartonellaspecies. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Chomel, B.B.; Kasten, R.W.; Henn, J.B.; Molia, S. Bartonella infection in domestic cats and wild felids. Ann. N. Y. Acad. Sci. 2006, 1078, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Leulmi, H.; Bitam, I.; Berenger, J.M.; Lepidi, H.; Rolain, J.M.; Almeras, L.; Raoult, D.; Parola, P. Competence of cimex lectularius bed bugs for the transmission of Bartonella quintana, the agent of trench fever. PLoS Negl. Trop. Dis. 2015, 9, e0003789. [Google Scholar]
- Kernif, T.; Leulmi, H.; Socolovschi, C.; Berenger, J.M.; Lepidi, H.; Bitam, I.; Rolain, J.M.; Raoult, D.; Parola, P. Acquisition and excretion of Bartonella quintana by the cat flea, ctenocephalides felis felis. Mol. Ecol. 2014, 23, 1204–1212. [Google Scholar] [CrossRef] [PubMed]
- Angelakis, E.; Socolovschi, C.; Raoult, D. Bartonella quintana in cimex hemipterus, rwanda. Am. J. Trop. Med. Hyg. 2013, 89, 986–987. [Google Scholar] [CrossRef] [PubMed]
- Doern, G.V. Detection of selected fastidious bacteria. Clin. Infect. Dis. 2000, 30, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Chamberlin, J.; Laughlin, L.; Gordon, S.; Romero, S.; Solorzano, N.; Regnery, R.L. Serodiagnosis of Bartonella bacilliformis infection by indirect fluorescence antibody assay: Test development and application to a population in an area of bartonellosis endemicity. J. Clin. Microbiol. 2000, 38, 4269–4271. [Google Scholar] [PubMed]
- Sanchez Clemente, N.; Ugarte-Gil, C.A.; Solorzano, N.; Maguina, C.; Pachas, P.; Blazes, D.; Bailey, R.; Mabey, D.; Moore, D. Bartonella bacilliformis: A systematic review of the literature to guide the research agenda for elimination. PLoS Negl. Trop. Dis. 2012, 6, e1819. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Xu, H.; Dai, H.; Meng, S.; Ye, C. Rapid and sensitive detection of listeria ivanovii by loop-mediated isothermal amplification of the smcl gene. PLoS ONE 2014, 9, e115868. [Google Scholar] [CrossRef] [PubMed]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liao, Y.; Ke, X.; Zhou, J.; Chen, Y.; Gao, L.; Chen, Q.; Yu, S. Comparison of reverse transcription loop-mediated isothermal amplification, conventional PCR and real-time PCR assays for japanese encephalitis virus. Mol. Biol. Rep. 2011, 38, 4063–4070. [Google Scholar] [CrossRef] [PubMed]
- Angkasekwinai, N.; Atkins, E.H.; Johnson, R.N.; Grieco, J.P.; Ching, W.M.; Chao, C.C. Rapid and sensitive detection of Bartonella bacilliformis in experimentally infected sand flies by loop-mediated isothermal amplification (LAMP) of the pap31 gene. PLoS Negl. Trop. Dis. 2014, 8, e3342. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Liberto, M.C.; Lamberti, A.G.; Marascio, N.; Matera, G.; Quirino, A.; Barreca, G.S.; Baudi, F.; Foca, A. Molecular identification of Bartonella quintana infection using species-specific real-time PCR targeting transcriptional regulatory protein (BQTR) gene. Mol. Cell. Probes 2011, 25, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Kabeya, H.; Yoshino, A.; Sekine, W.; Suzuki, K.; Tamate, H.B.; Yamazaki, S.; Chomel, B.B.; Maruyama, S. Japanese macaques (macaca fuscata) as natural reservoir of Bartonella quintana. Emerg. Infect. Dis. 2015, 21, 2168–2170. [Google Scholar] [CrossRef] [PubMed]
- Zeaiter, Z.; Liang, Z.; Raoult, D. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsz gene sequences. J. Clin. Microbiol. 2002, 40, 3641–3647. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, E.M.; Perez Vera, C.; Pulliainen, A.T.; Sironen, T.; Aaltonen, K.; Kortet, R.; Harkonen, L.; Harkonen, S.; Paakkonen, T.; Nieminen, P.; et al. Molecular detection of Bartonella spp. In deer ked pupae, adult keds and moose blood in finland. Epidemiol. Infect. 2015, 143, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Kim, E.J.; Kilgore, P.E.; Kim, S.A.; Takahashi, H.; Ohnishi, M.; Anh, D.D.; Dong, B.Q.; Kim, J.S.; Tomono, J.; et al. Clinical evaluation of a loop-mediated isothermal amplification (LAMP) assay for rapid detection of neisseria meningitidis in cerebrospinal fluid. PLoS ONE 2015, 10, e0122922. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zhang, L.; Fan, D.; Zhang, X.; Liu, H.; Lu, Q.; Xu, Q. Rapid, simple and sensitive detection of q fever by loop-mediated isothermal amplification of the htpab gene. PLoS Negl. Trop. Dis. 2013, 7, e2231. [Google Scholar] [CrossRef] [PubMed]
- Paris, D.H.; Blacksell, S.D.; Newton, P.N.; Day, N.P. Simple, rapid and sensitive detection of orientia tsutsugamushi by loop-isothermal DNA amplification. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Iseki, H.; Alhassan, A.; Ohta, N.; Thekisoe, O.M.; Yokoyama, N.; Inoue, N.; Nambota, A.; Yasuda, J.; Igarashi, I. Development of a multiplex loop-mediated isothermal amplification (MLAMP) method for the simultaneous detection of bovine babesia parasites. J. Microbiol. Methods 2007, 71, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Lau, Y.L.; Fong, M.Y.; Mahmud, R.; Chang, P.Y.; Palaeya, V.; Cheong, F.W.; Chin, L.C.; Anthony, C.N.; Al-Mekhlafi, A.M.; Chen, Y. Specific, sensitive and rapid detection of human plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar. J. 2011, 10, 197. [Google Scholar] [CrossRef] [PubMed]
- Minnick, M.F.; Smitherman, L.S.; Samuels, D.S. Mitogenic effect of Bartonella bacilliformis on human vascular endothelial cells and involvement of groel. Infect. Immun. 2003, 71, 6933–6942. [Google Scholar] [CrossRef] [PubMed]
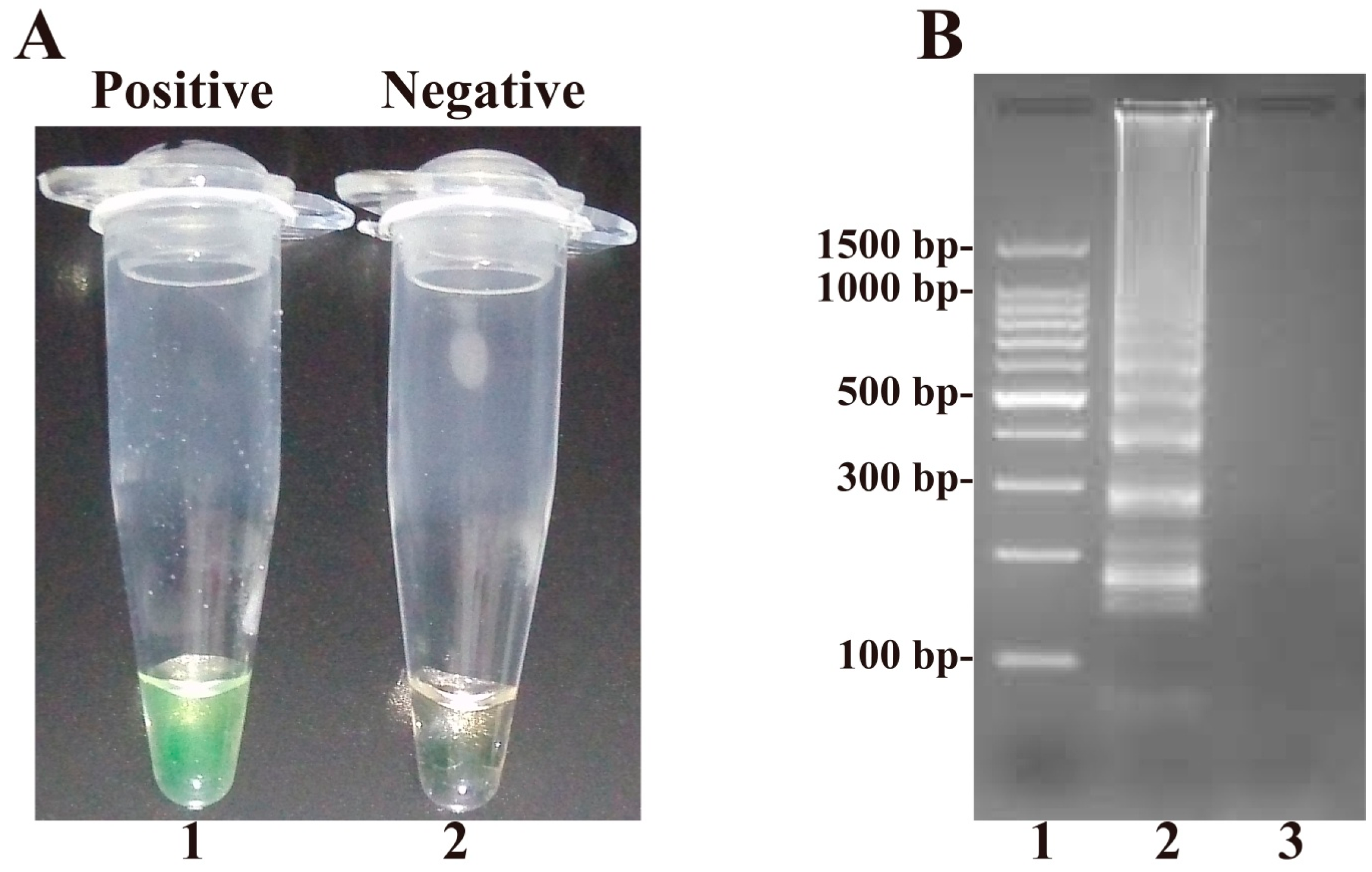
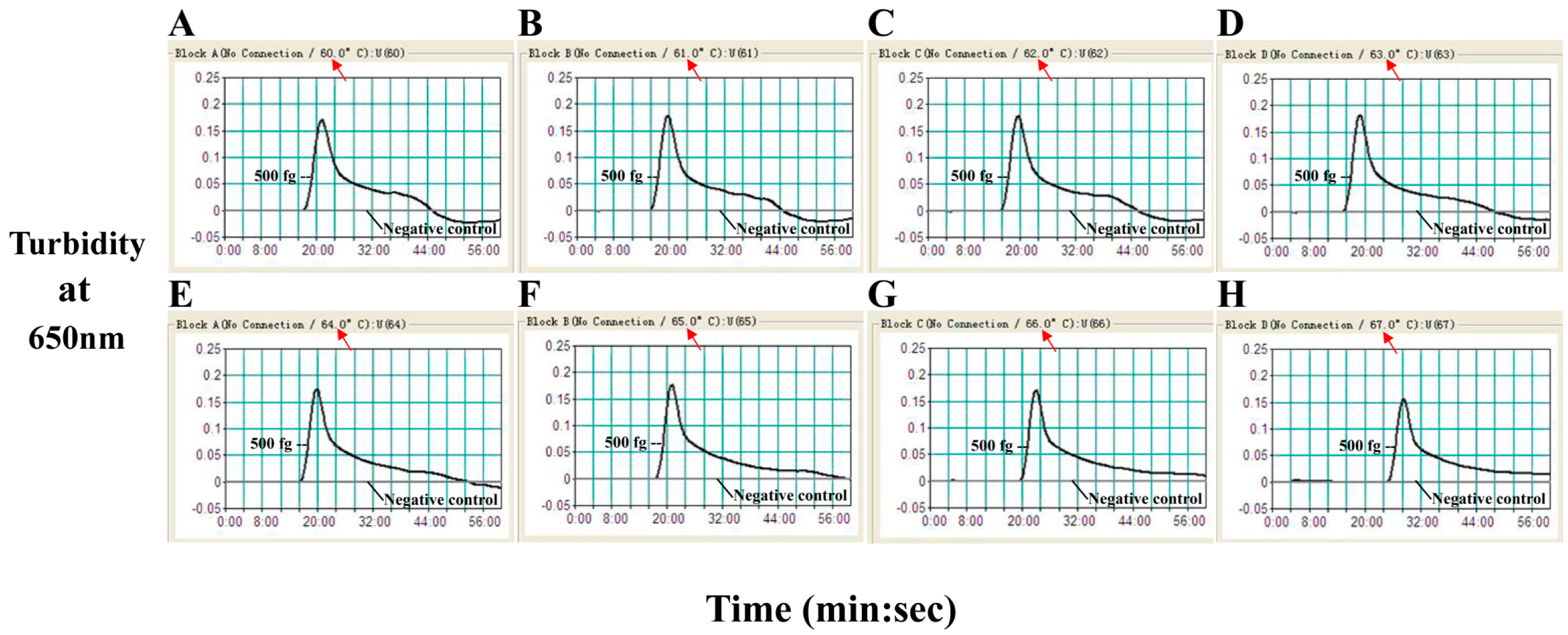
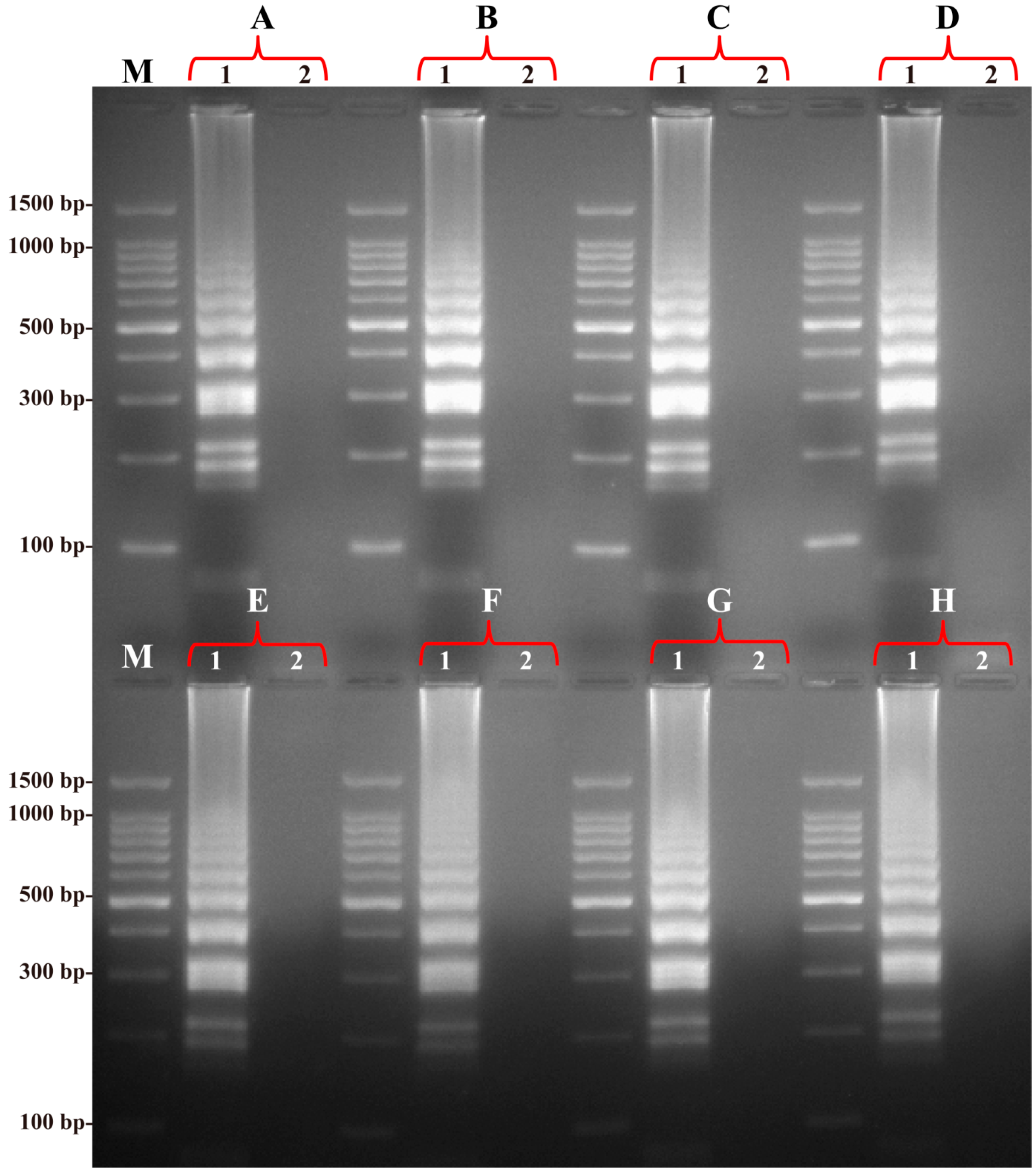


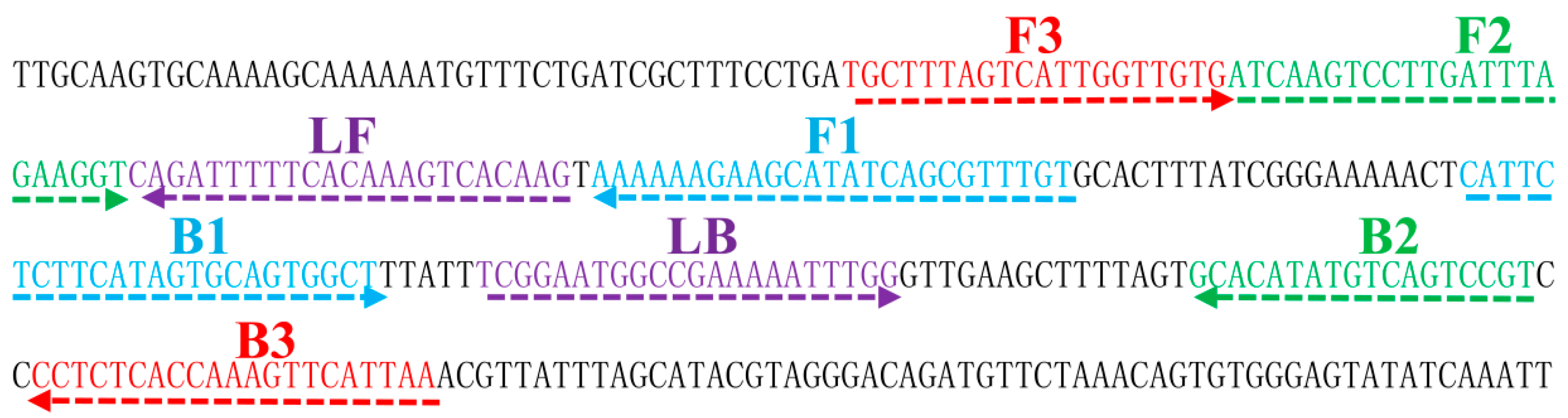
| Assay | Regions Recognized | LOD (DNA/Reaction) | Fastest Time (min) |
|---|---|---|---|
| LAMP | 8 | 125 fg | 18 |
| qPCR | 3 | 500 fg | 35 |
| Positive Rate (%) (No. of Positives/Total No. of Samples) | ||
|---|---|---|
| Samples | LAMP | qPCR |
| 100 rhesus | 22.0 (22/100) | 8.0 (8/100) |
| 20 rhesus feeders | 20.0 (4/20) | 5.0 (1/20) |
| Bacterium | Source of Strain | No. of Strains |
|---|---|---|
| B. quintana | str. Toulouse | 1 |
| B. quintana | isolated strains (ICDC13001-13009) | 9 |
| B. henselae | isolated strains (ICDC14112) | 1 |
| B. elizabethae | isolated strains (ICDC14116) | 1 |
| B. alsatica | isolated strains (ICDC14117) | 1 |
| B. koehlerae | isolated strains (ICDC15002) | 1 |
| B. vinsonii subsp. berkhoffii | isolated strains (ICDC13010) | 1 |
| B. vinsonii subsp. vinsonii | isolated strains (ICDC10001) | 1 |
| B. vinsonii subsp. arupensis | isolated strains (ICDC10002) | 1 |
| B. tribocorum | isolated strains (ICDC11013) | 1 |
| B. grahamii | isolated strains (ICDC10161) | 1 |
| B. clarridgeiae | isolated strains (ICDC10181) | 1 |
| B. bacilliformis | isolated strains (ICDC13144) | 1 |
| B. doshiae | isolated strains (ICDC13167) | 1 |
| B. mayotimonensis | isolated strains (ICDC10009) | 1 |
| Yersinia enterocolitica | ATCC 23715 | 1 |
| Pseudomonas aeruginosa | ATCC 15442 | 1 |
| Enterococcus faecalis | ATCC 35667 | 1 |
| Aeromonas hydrophila | ATCC 7966 | 1 |
| Enterobacter sakazakii | ATCC 51329 | 1 |
| Campylobacter jejuni | ATCC 33291 | 1 |
| Bacillus cereus | Isolated strains (ICDC10118) | 1 |
| Salmonella typhimurium | Isolated strains (ICDC12113) | 1 |
| Vibrio cholera | Isolated strains (ICDC09111) | 1 |
| Escherichia coli | Isolated strains (ICDC08117) | 1 |
| Listeria monocytogenes | Isolated strains (ICDC12211) | 1 |
| Shigella sonnei | ATCC9372 | 1 |
| Staphylococcus aureus | ATCC25923 | 1 |
| Assay | Primer | Sequence (5′–3′) | Length (nt) |
|---|---|---|---|
| groEL_LAMP | F3 | TGCTTTAGTCATTGGTTGTG | 20 |
| B3 | TTAATGAACTTTGGTGAGAGG | 21 | |
| FIP | ACAAACGCTGATATGCTTCTTTTTT-ATCAAGTCCTTGATTTAGAAGGT | 48 | |
| BIP | CATTCTCTTCATAGTGCAGTGGCT-ACGGACTGACATATGTGC | 42 | |
| LF | CTTGTGACTTTGTGAAAAATCTG | 23 | |
| LB | TCGGAATGGCCGAAAAATTTGG | 22 | |
| groEL_qPCR | F | TGCCAAGTATGCGAGTTCCTG | 21 |
| Probe | Cy5-TCTGCGCCCGGTTCTGAAATGCCT-BHQ2 | 24 | |
| R | TCAGAGTTAGGTGCAAGTTCTATGG | 25 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Niu, L.; Luo, L.; Song, X.; Sun, J.; Liu, Q. Rapid, Sensitive Detection of Bartonella quintana by Loop-Mediated Isothermal Amplification of the groEL Gene. Int. J. Mol. Sci. 2016, 17, 1902. https://doi.org/10.3390/ijms17121902
Hu S, Niu L, Luo L, Song X, Sun J, Liu Q. Rapid, Sensitive Detection of Bartonella quintana by Loop-Mediated Isothermal Amplification of the groEL Gene. International Journal of Molecular Sciences. 2016; 17(12):1902. https://doi.org/10.3390/ijms17121902
Chicago/Turabian StyleHu, Shoukui, Lina Niu, Lijuan Luo, Xiuping Song, Jimin Sun, and Qiyong Liu. 2016. "Rapid, Sensitive Detection of Bartonella quintana by Loop-Mediated Isothermal Amplification of the groEL Gene" International Journal of Molecular Sciences 17, no. 12: 1902. https://doi.org/10.3390/ijms17121902





