Immune-Related Gene Expression Patterns in GPV- or H9N2-Infected Goose Spleens
Abstract
:1. Introduction
2. Results
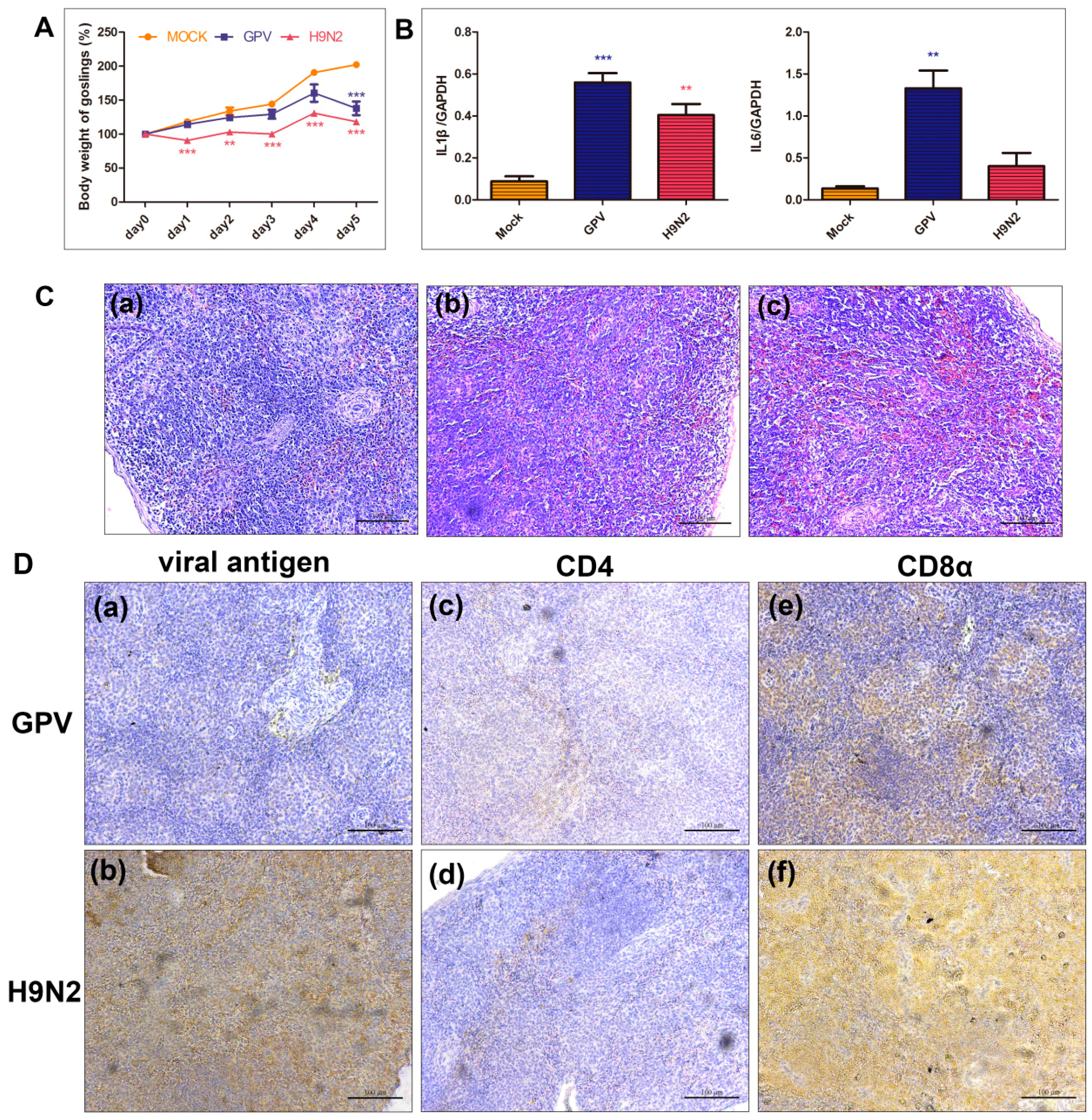
2.1. Global Characteristics in Response to GPV and H9N2
2.2. Transcriptional Data Profile of the Virus-Infected Goslings
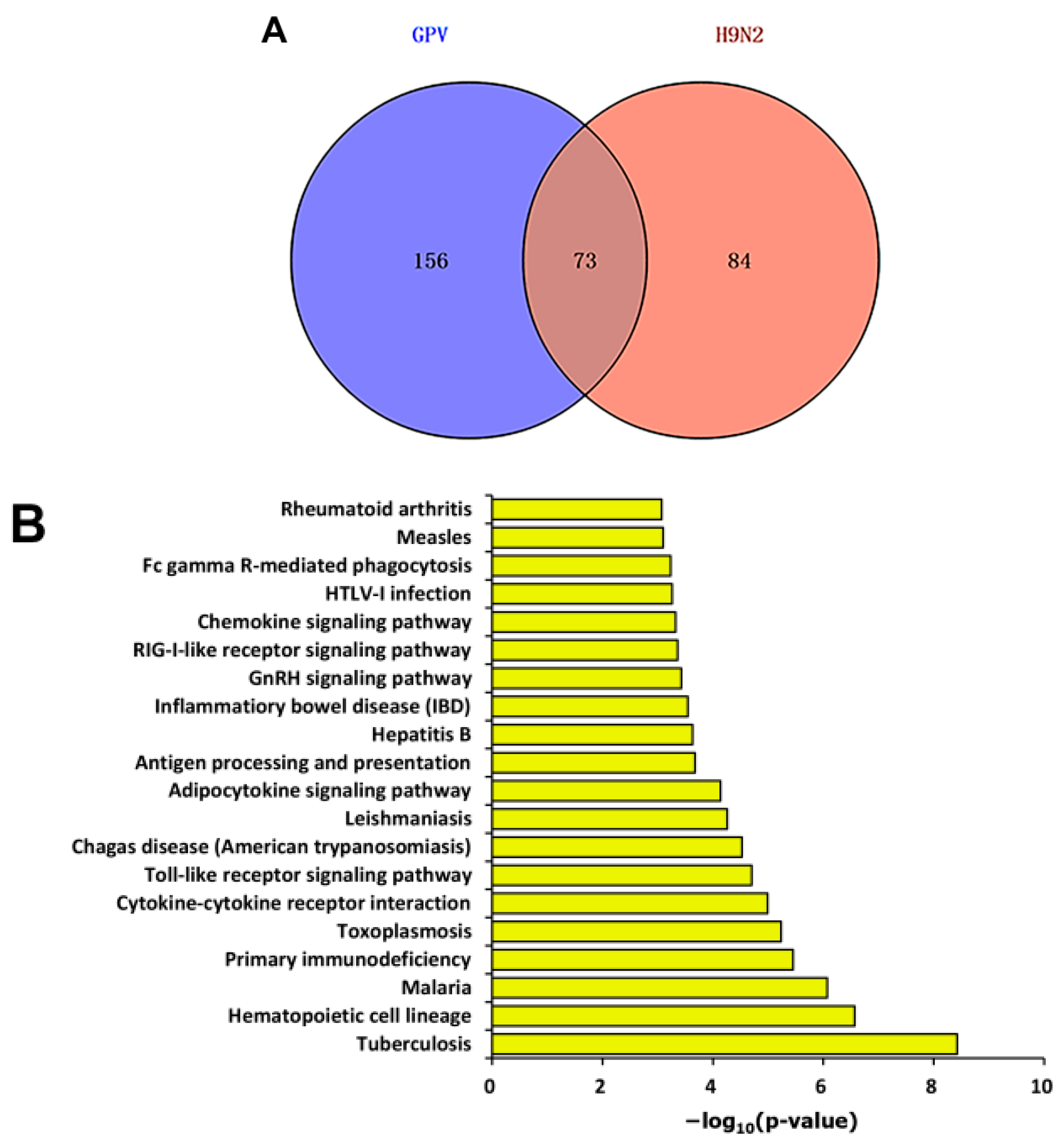
2.3. Differential Expression Pattern after Infection
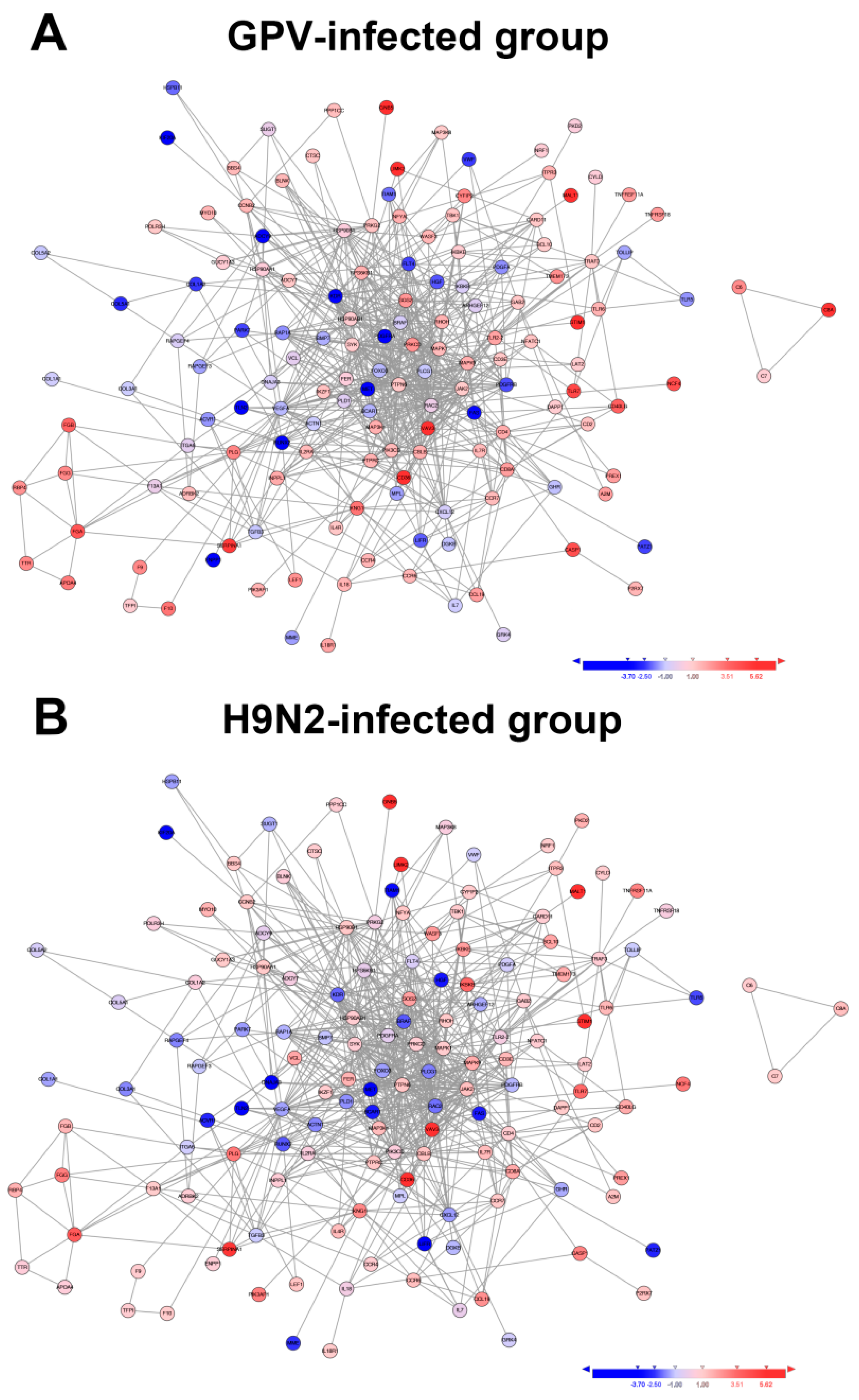
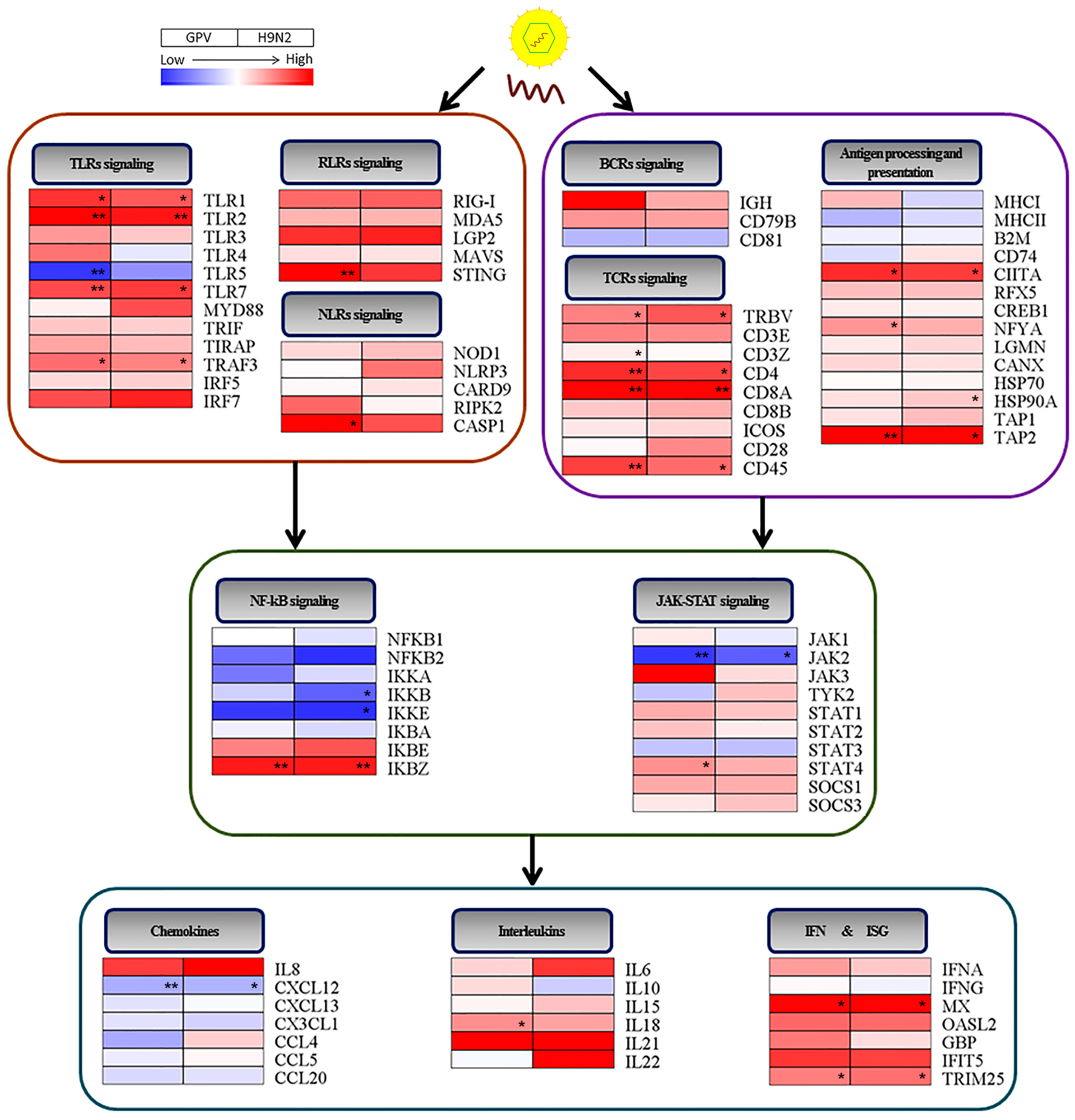
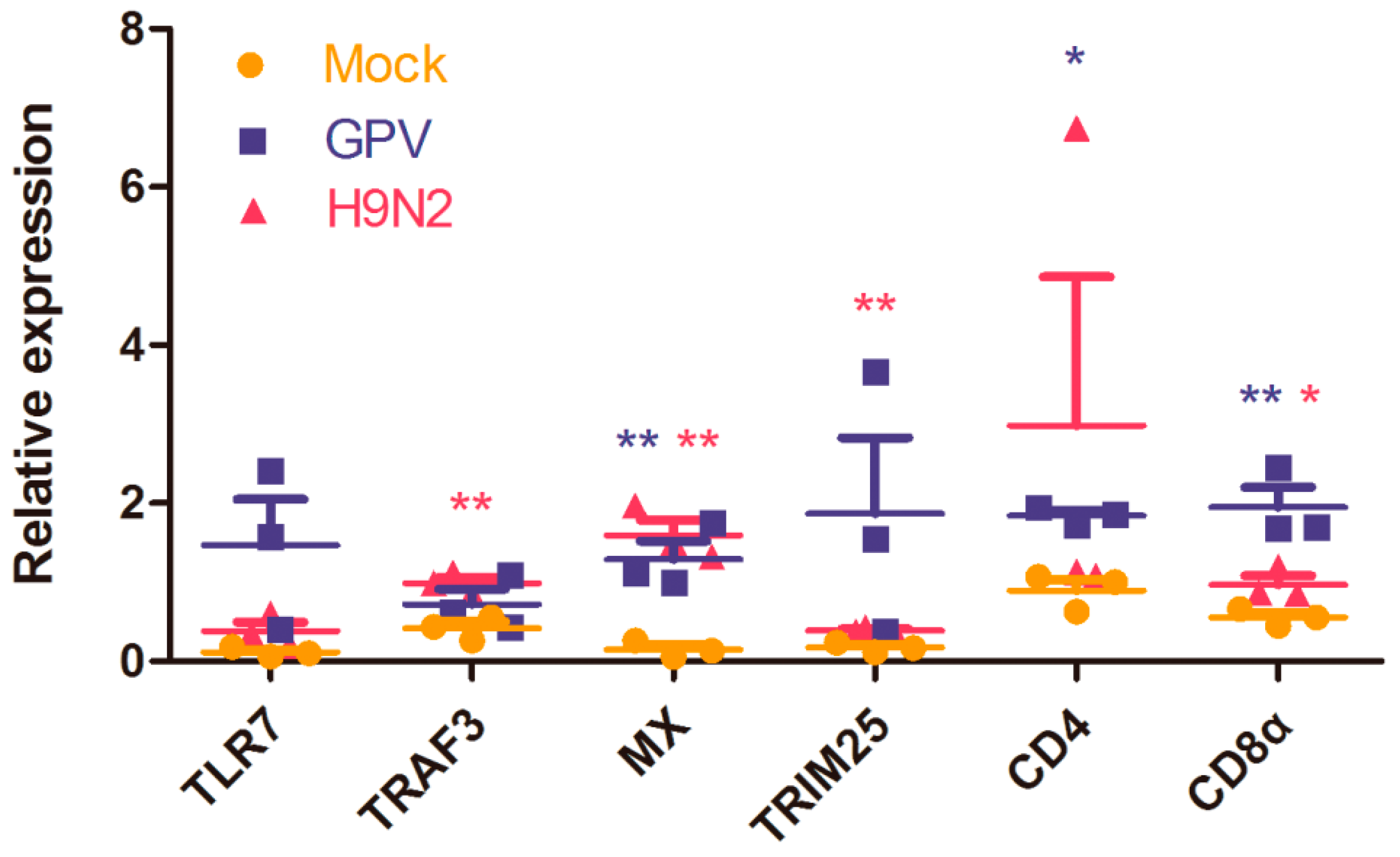
2.4. Dynamics of Immune Relevant Genes during the Defense Response
2.5. Innate Immune and Adaptive Immune Response after Infection with the Two Viruses
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Animal Experiment and Spleen Tissue Collection
4.3. H & E Staining and Immunohistochemical (IHC) Analysis
4.4. Total RNA Isolation and Illumina Sequencing
4.5. Transcriptome Data Analysis
4.6. RT-qPCR Analysis
4.7. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Xu, X.; Subbarao, K.; Cox, N.J.; Guo, Y. Genetic characterization of the pathogenic influenza a/goose/guangdong/1/96 (H5N1) virus: Similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 1999, 261, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Webster, R.G.; Guan, Y.; Peiris, M.; Walker, D.; Krauss, S.; Zhou, N.N.; Govorkova, E.A.; Ellis, T.M.; Dyrting, K.; Sit, T. Characterization of H5N1 influenza viruses that continue to circulate in geese in southeastern China. J. Virol. 2002, 76, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.E.; Green, S.W.; Young, N.S. Goose parvovirus—An autonomous member of the dependovirus genus? Virology 1995, 210, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Dressy, D. A viral disease of goslings. Acta Vet. Acad. Sci. Hung. 1967, 17, 443–448. [Google Scholar]
- Chen, H.; Dou, Y.; Tang, Y.; Zhang, Z.; Zheng, X.; Niu, X.; Yang, J.; Yu, X.; Diao, Y. Isolation and genomic characterization of a duck-origin GPV-related parvovirus from cherry valley ducklings in China. PLoS ONE 2015, 10, e0140284. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.N.; Krauss, S.; Webster, R.G. H9N2 influenza a viruses from poultry in Asia have human virus-like receptor specificity. Virology 2001, 281, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Shortridge, K.F.; Krauss, S.; Webster, R.G. Molecular characterization of H9N2 influenza viruses: Were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 1999, 96, 9363–9367. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Chen, S.; Yan, B.; Chen, H.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Liu, F.; Yang, Q. LPAIV H9N2 drives the differential expression of goose interferons and proinflammatory cytokines in both in vitro and in vivo studies. Front. Microbiol. 2016, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Hui, R.K.; Leung, F.C. Differential expression profile of chicken embryo fibroblast DF-1 cells infected with cell-adapted infectious bursal disease virus. PLoS ONE 2015, 10, e0111771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, J.; Bao, Y.; Ye, J.; Shao, H.; Qian, K.; Qin, A. Transcriptional profiling of host gene expression in chicken embryo fibroblasts infected with reticuloendotheliosis virus strain HA1101. PLoS ONE 2015, 10, e0126992. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Simonds, W.F.; Zhang, J.-H. New dimensions in G protein signalling: G β5 and the RGS proteins. Pharm. Acta Helv. 2000, 74, 333–336. [Google Scholar] [CrossRef]
- Bustelo, X.R. Regulatory and signaling properties of the Vav family. Mol. Cell. Biol. 2000, 20, 1461–1477. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Deng, L.; Ea, C.-K.; Xia, Z.-P.; Chen, Z.J. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 2004, 14, 289–301. [Google Scholar] [CrossRef]
- Thome, M. Carma1, BCL-10 and MALT1 in lymphocyte development and activation. Nat. Rev. Immunol. 2004, 4, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.-M.; Gale, M.; Akira, S. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Burns, K.; Tschopp, J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Harty, J.T.; Tvinnereim, A.R.; White, D.W. CD8+ T cell effector mechanisms in resistance to infection. Annu Rev. Immunol. 2000, 18, 275–308. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.K.; Lichtman, A.H.; Pillai, S. Cellular and Molecular Immunology, 8th ed.; Elsevier Health Sciences: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Brasier, A.R. The NF-κB regulatory network. Cardiovasc. Toxicol. 2006, 6, 111–130. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef] [PubMed]
- Staeheli, P.; Puehler, F.; Schneider, K.; Göbel, T.W.; Kaspers, B. Cytokines of birds: Conserved functions-a largely different look. J. Interferon Cytokine Res. 2001, 21, 993–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheller, J.; Chalaris, A.; Schmidt-Arras, D.; Rose-John, S. The pro-and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 2011, 1813, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.J.; Williams, B. Signal transduction in the interferon system. Semin. Oncol. 1998, 25, 14–22. [Google Scholar] [PubMed]
- De Veer, M.J.; Holko, M.; Frevel, M.; Walker, E.; Der, S.; Paranjape, J.M.; Silverman, R.H.; Williams, B.R. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001, 69, 912–920. [Google Scholar] [PubMed]
- Von Mende, N.; Bird, D.M.; Albert, P.S.; Riddle, D.L. Dpy-13: A nematode collagen gene that affects body shape. Cell 1988, 55, 567–576. [Google Scholar] [CrossRef]
- Hortsch, M.; Goodman, C.S. Cell and substrate adhesion molecules in drosophila. Annu. Rev. Cell Biol. 1991, 7, 505–557. [Google Scholar] [CrossRef] [PubMed]
- Thoma-Uszynski, S.; Stenger, S.; Takeuchi, O.; Ochoa, M.T.; Engele, M.; Sieling, P.A.; Barnes, P.F.; Röllinghoff, M.; Bölcskei, P.L.; Wagner, M. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science 2001, 291, 1544–1547. [Google Scholar] [CrossRef] [PubMed]
- Echchannaoui, H.; Frei, K.; Schnell, C.; Leib, S.L.; Zimmerli, W.; Landmann, R. Toll-like receptor 2—Deficient mice are highly susceptible to streptococcus pneumoniae meningitis because of reduced bacterial clearing and enhanced inflammation. J. Infect. Dis. 2002, 186, 798–806. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.; Sareneva, T.; Julkunen, I.; Matikainen, S. IFNs activate toll-like receptor gene expression in viral infections. Genes Immun. 2001, 2, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Sousa, C.R. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Chen, D.; Xiong, M.; Zhu, J.; Lin, X.; Wang, L.; Zhang, J.; Chen, L.; Zhang, H.; Chen, H. Insights into the increasing virulence of the swine-origin pandemic H1N1/2009 influenza virus. Sci. Rep. 2013, 3, 132. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Weber, F.; Wagner, V.; Rasmussen, S.B.; Hartmann, R.; Paludan, S.R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006, 80, 5059–5064. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Ma, Z.; Barber, G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 2009, 461, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Rothenfusser, S.; Goutagny, N.; DiPerna, G.; Gong, M.; Monks, B.G.; Schoenemeyer, A.; Yamamoto, M.; Akira, S.; Fitzgerald, K.A. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 2005, 175, 5260–5268. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Hirai, R.; Loo, Y.-M.; Owen, D.; Johnson, C.L.; Sinha, S.C.; Akira, S.; Fujita, T.; Gale, M. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 2007, 104, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, T.-D.; Lamkanfi, M.; Núñez, G. Intracellular NOD-like receptors in host defense and disease. Immunity 2007, 27, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.G.; Dash, P.; Aldridge, J.R.; Ellebedy, A.H.; Reynolds, C.; Funk, A.J.; Martin, W.J.; Lamkanfi, M.; Webby, R.J.; Boyd, K.L. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 2009, 30, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Kochs, G.; Haller, O. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc. Natl. Acad. Sci. USA 1999, 96, 2082–2086. [Google Scholar] [CrossRef] [PubMed]
- Turan, K.; Mibayashi, M.; Sugiyama, K.; Saito, S.; Numajiri, A.; Nagata, K. Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic Acids Res. 2004, 32, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Gack, M.U.; Shin, Y.C.; Joo, C.-H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef] [PubMed]
- Aaronson, D.S.; Horvath, C.M. A road map for those who don’t know JAK-STAT. Science 2002, 296, 1653–1655. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo, C.R., Jr. Modulation of major histocompatibility complex antigen expression by viral infection. Am. J. Pathol. 1994, 144, 637. [Google Scholar] [PubMed]
- Xing, Z.; Cardona, C.J.; Li, J.; Dao, N.; Tran, T.; Andrada, J. Modulation of the immune responses in chickens by low-pathogenicity avian influenza virus H9N2. J. Gen. Virol. 2008, 89, 1288–1299. [Google Scholar] [CrossRef] [PubMed]
- Abendroth, A.; Slobedman, B.; Lee, E.; Mellins, E.; Wallace, M.; Arvin, A.M. Modulation of major histocompatibility class II protein expression by varicella-zoster virus. J. Virol. 2000, 74, 1900–1907. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D. Introduction to NF-κB: Players, pathways, perspectives. Oncogene 2006, 25, 6680–6684. [Google Scholar] [CrossRef] [PubMed]
- Hiscott, J.; Nguyen, T.A.; Arguello, M.; Nakhaei, P.; Paz, S. Manipulation of the nuclear factor-κB pathway and the innate immune response by viruses. Oncogene 2006, 25, 6844–6867. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-Y.; Lee, J.-S.; Ko, Y.-G.; Kim, J.-I.; Seo, J.-S. Heat shock protein 70 inhibits apoptosis downstream of cytochrome c release and upstream of caspase-3 activation. J. Biol. Chem. 2000, 275, 25665–25671. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Q.; Cheng, B.; Yan, B.; Yan, X.; Zhao, Q.; Wang, M.; Jia, R.; Zhu, D.; Liu, M. Age-related development and tissue distribution of T cell markers (CD4 and CD8a) in Chinese goose. Immunobiology 2015, 220, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome without a genome from RNA-seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B. TIGR Gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Liu, F.; Chen, S.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Sun, K.; Wu, Y.; Chen, X. Transcriptome analysis and identification of differentially expressed transcripts of immune-related genes in spleen of gosling and adult goose. Int. J. Mol. Sci. 2015, 16, 22904–22926. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| Sample | RINs | Raw Reads | Clean Reads | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|
| Mock-1 | 9.9 | 55,922,366 | 55,647,608 | 94.02 | 47.00% |
| Mock-2 | 9.9 | 45,717,764 | 45,490,282 | 93.94 | 47.50% |
| Mock-3 | 10.0 | 49,752,976 | 49,480,934 | 94.09 | 47.00% |
| GPV-1 | 10.0 | 54,915,460 | 54,607,164 | 93.77 | 47.00% |
| GPV-2 | 9.9 | 50,402,808 | 50,120,838 | 93.78 | 47.00% |
| GPV-3 | 9.9 | 43,356,716 | 43,137,380 | 94.03 | 47.00% |
| H9N2-1 | 9.9 | 49,921,838 | 49,641,084 | 94.03 | 46.00% |
| H9N2-2 | 9.8 | 50,382,454 | 50,133,860 | 94.17 | 47.00% |
| H9N2-3 | 9.9 | 54,094,468 | 53,823,010 | 94.00 | 47.50% |
| Gene Symbol | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Size/bp |
|---|---|---|---|
| IL-1β | TCCGCCAGCCGCAAAGTG | CGCTCATCACGCAGGACA | 136 |
| IL-6 | GCTTTGTGAGGAGGGATT | CCGTTAGACACTGGGGTT | 120 |
| CD8α | AGAGACGAGCAAGGAGAA | GACCAGGGCAATGAGAAG | 97 |
| CD4 | TTTCAACGCCACAGCAGA | GTGCCTCAACTGGATTTT | 127 |
| Mx | TTCACAGCAATGGAAAGGGA | ATTAGTGTCGGGTCTGGGA | 183 |
| TRIM25 | CCACCACCCTCAGCGTTTC | GCCATAGCAGATGCCAAT | 127 |
| TAP2 | TCTTCCAGCAGACCACAGC | AAGGGGCACCTCAAGCAG | 188 |
| TLR7 | CACAGAAAAATGGTACCTC | TACATCGCAGGGTAAACT | 117 |
| GAPDH | CATCTTCCAGGAGCGCGACC | AGACACCGGTGGACTCCACA | 80 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Wang, A.; Sun, L.; Liu, F.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Yang, Q.; Wu, Y.; et al. Immune-Related Gene Expression Patterns in GPV- or H9N2-Infected Goose Spleens. Int. J. Mol. Sci. 2016, 17, 1990. https://doi.org/10.3390/ijms17121990
Chen S, Wang A, Sun L, Liu F, Wang M, Jia R, Zhu D, Liu M, Yang Q, Wu Y, et al. Immune-Related Gene Expression Patterns in GPV- or H9N2-Infected Goose Spleens. International Journal of Molecular Sciences. 2016; 17(12):1990. https://doi.org/10.3390/ijms17121990
Chicago/Turabian StyleChen, Shun, Anqi Wang, Lipei Sun, Fei Liu, Mingshu Wang, Renyong Jia, Dekang Zhu, Mafeng Liu, Qiao Yang, Ying Wu, and et al. 2016. "Immune-Related Gene Expression Patterns in GPV- or H9N2-Infected Goose Spleens" International Journal of Molecular Sciences 17, no. 12: 1990. https://doi.org/10.3390/ijms17121990