Cytokinins and Expression of SWEET, SUT, CWINV and AAP Genes Increase as Pea Seeds Germinate
Abstract
:1. Introduction
2. Results
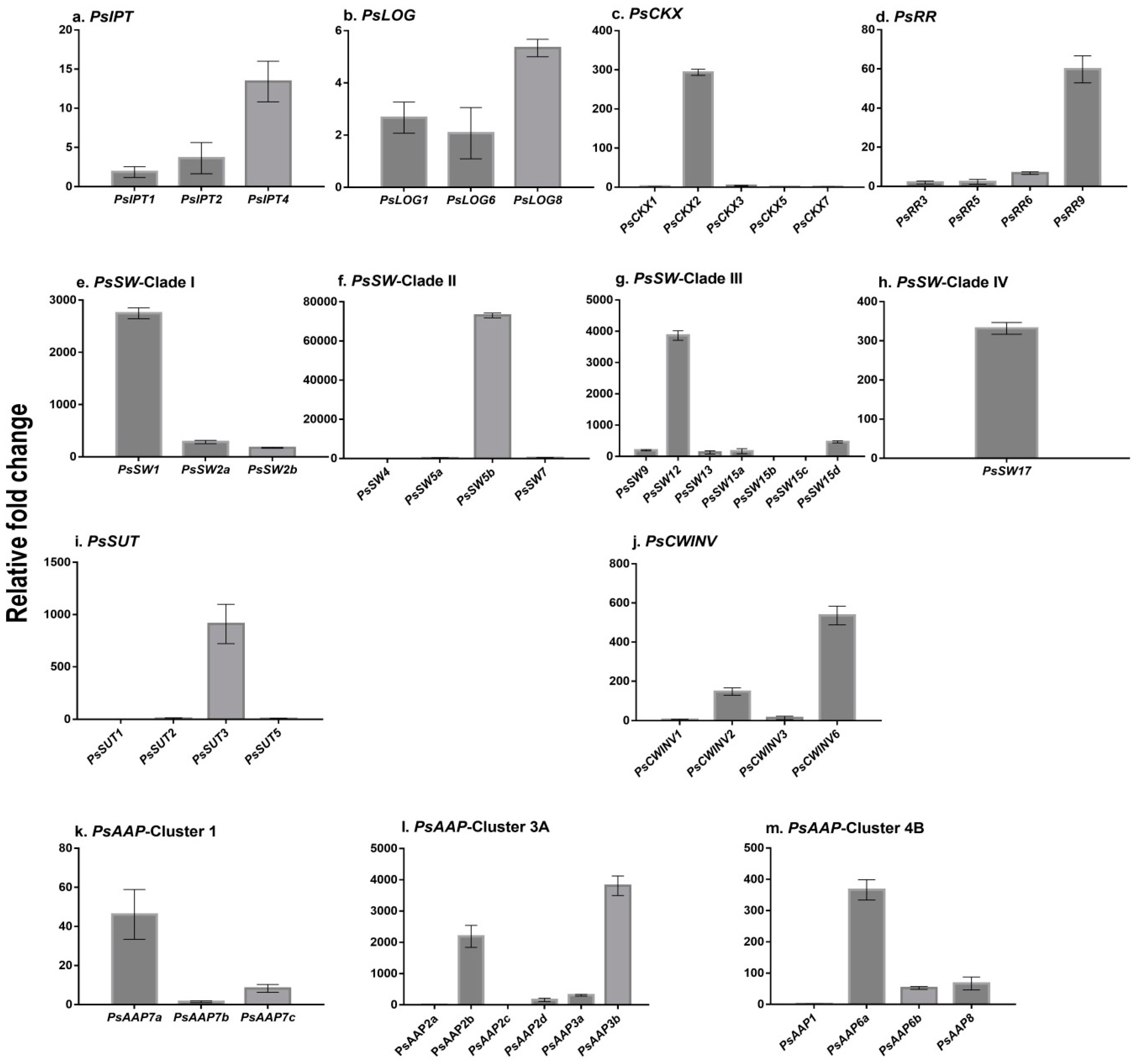
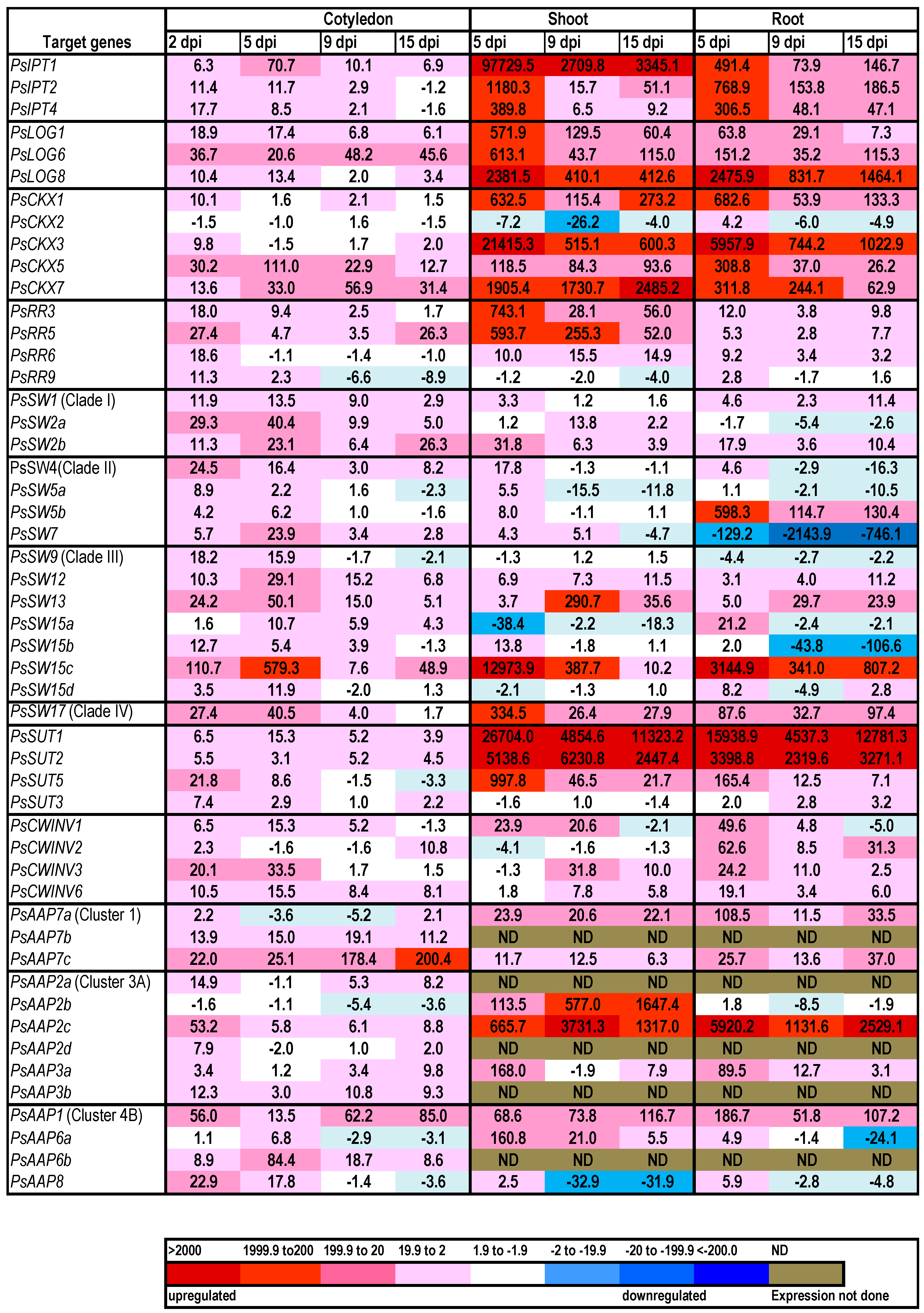
2.1. Cytokinin Biosynthesis and Metabolism in the Germinating Pea
2.2. Expression of Transporter Genes in the Germinating Seed
3. Discussion
4. Materials and Methods
4.1. Plant Material and Sample Preparation
4.2. RNA Isolation and Target Gene Isolation
4.3. Real-Time Reverse Transcription Quantitative PCR (RT-qPCR)
4.4. Cytokinin Analyses
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yu, S.-M.; Lo, S.-F.; Ho, T.-H.D. Source-sink communication: Regulated by hormone, nutrient, and stress cross-signaling. Trends Plant Sci. 2015, 20, 844–857. [Google Scholar] [CrossRef] [PubMed]
- Roche, J.; Love, J.; Guo, Q.; Song, J.; Cao, M.; Fraser, K.; Huege, J.; Jones, C.; Novák, O.; Turnbull, M.H.; et al. Metabolic changes and associated cytokinin signals in response to nitrate assimilation in roots and shoots of Lolium perenne. Physiol. Plant. 2016, 156, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ruan, Y.-L. Shoot–root carbon allocation, sugar signaling and their coupling with nitrogen uptake and assimilation. Funct. Plant Biol. 2016, 43, 105–113. [Google Scholar] [CrossRef]
- Braun, D.M.; Wang, L.; Ruan, Y.-L. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J. Exp. Bot. 2014, 65, 1713–1735. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.-L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M. Transporters for amino acids in plant cells: Some functions and many unknowns. Curr. Opin. Plant Biol. 2012, 15, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.P.; Tegeder, M. Connecting source with sink: The role of Arabidopsis AAP8 in phloem loading of amino acids. Plant Physiol. 2016, 171, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Patrick, J.W.; Offler, C.E. Compartmentation of transport and transfer events in developing seeds. J. Exp. Bot. 2001, 52, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Rosche, E.; Blackmore, D.; Tegeder, M.; Richardson, T.; Schroeder, H.; Higgins, T.J.V.; Frommer, W.B.; Offler, C.E.; Patrick, J.W. Seed-specific overexpression of a potato sucrose transporter increases sucrose uptake and growth rates of developing cotyledons. Plant J. 2002, 30, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M. Transporters involved in source to sink partitioning of amino acids and ureides: Opportunities for crop improvement. J. Exp. Bot. 2014, 65, 1865–1878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Garneau, M.G.; Majumdar, R.; Grant, J.; Tegeder, M. Improvement of pea biomass and seed productivity by simultaneous increase of phloem and embryo loading with amino acids. Plant J. 2015, 81, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Eom, J.S.; Chen, L.-Q.; Sosso, D.; Benjamin, T.J.; Lin, I.W.; Qu, X.-Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M.; Ward, J.M. Molecular evolution of plant AAP and LHT amino acid transporters. Front. Plant Sci. 2012, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Beevers, L.; Guernsey, F.S. Changes in some nitrogenous components during the germination of pea seeds. Plant Physiol. 1966, 41, 1455–1458. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, T.M.; Nesi, A.N.; Araújo, W.L.; Braun, H.-P. Amino acid catabolism in plants. Mol. Plant 2015, 8, 1563–1579. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.R.; Reinders, A.; Ward, J.M. Transport function of rice amino acid permeases (AAPs). Plant Cell Physiol. 2015, 56, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Dhandapani, P.; Song, J.; Novak, O.; Jameson, P.E. Infection by Rhodococcus fascians maintains cotyledons as a sink tissue for the pathogen. Ann. Bot. 2016. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Gu, X.; Xue, L.-J.; Leebens-Mack, J.H.; Tsai, C.-J. Bayesian phylogeny of sucrose transporters: Ancient origins, differential expansion and convergent evolution in monocots and dicots. Front. Plant Sci. 2014, 5, 615. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Qu, H.; Dibley, K.E.; Offler, C.E. A suite of sucrose transporters expressed in coats of developing legume seeds includes novel pH-independent facilitators. Plant J. 2007, 49, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Reinders, A.; Sivitz, A.B.; Ward, J.M. Evolution of plant sucrose uptake transporters. Front. Plant Sci. 2012, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Scofield, G.N.; Aoki, N.; Hirose, T.; Takano, M.; Jenkins, C.L.; Furbank, R.T. The role of the sucrose transporter, OsSUT1, in germination and early seedling growth and development of rice plants. J. Exp. Bot. 2007, 58, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.; Hsiao, H.-H.; Chen, H.-J.; Chang, C.-W.; Wang, S.-J. Influence of temperature on the expression of the rice sucrose transporter 4 gene, OsSUT4, in germinating embryos and maturing pollen. Acta Physiol. Plant. 2014, 36, 217–229. [Google Scholar] [CrossRef]
- Slewinski, T.L.; Meeley, R.; Braun, D.M. Sucrose transporter 1 functions in phloem loading in maize leaves. J. Exp. Bot. 2009, 60, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Bick, J.A.; Neelam, A.; Smith, E.; Nelson, S.J.; Hall, J.L.; Williams, L.E. Expression analysis of a sucrose carrier in the germinating seedling of Ricinus communis. Plant Mol. Biol. 1998, 38, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Tegeder, M.; Wang, X.D.; Frommer, W.B.; Offler, C.E.; Patrick, J.W. Sucrose transport into developing seeds of Pisum sativum L. Plant J. 1999, 18, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Borisjuk, L.; Wobus, U. Sugar import and metabolism during seed development. Trends Plant Sci. 1997, 2, 169–174. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-Y.; Huh, J.-H.; Yu, Y.-C.; Ho, L.-H.; Chen, L.-Q.; Tholl, D.; Frommer, W.B.; Guo, W.-J. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 2015, 83, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- Chandran, D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance. IUBMB Life 2015, 67, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef] [PubMed]
- Klemens, P.A.W.; Patzke, K.; Deitmer, J.; Spinner, L.; Le Hir, R.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Neuhaus, H.E. Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth and stress tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Yang, C.; Tian, Z.; Li, J. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.-L.; Jin, Y.; Yang, Y.-J.; Li, G.-J.; Boyer, J.S. Sugar input, metabolism, and signaling mediated by invertase: Roles in development, yield potential, and response to drought and heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Roitsch, T.; Gonzalez, M.C. Function and regulation of plant invertases: sweet sensations. Trends Plant Sci. 2004, 9, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Balibrea Lara, M.E.; Gonzalez Garcia, M.-C.; Fatima, T.; Ehneß, R.; Lee, T.K.; Proels, R.; Tanner, W.; Roitsch, T. Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 2004, 16, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Sheen, J.; Muller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.H.; Taliercio, E.W.; Chourey, P.S. The Miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell. 1996, 8, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Ehneß, R.; Roitsch, T. Co-ordinated induction of mRNAs for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. Plant J. 1997, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Rijavec, T.; Kovač, M.; Kladnik, A.; Chourey, P.S.; Dermastia, M. A comparative study on the role of cytokinins in caryopsis development in the maize miniature1 seed mutant and its wild type. J. Integr. Plant Biol. 2009, 51, 840–849. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Jiang, L.; Jameson, P.E. Expression patterns of Brassica napus genes implicate IPT, CKX, sucrose transporter, cell wall invertase and amino acid permease gene family members in leaf, flower, silique and seed development. J. Exp. Bot. 2015, 66, 5067–5082. [Google Scholar] [CrossRef] [PubMed]
- Jameson, P.E.; Song, J. Cytokinin: A key driver of seed yield. J. Exp. Bot. 2016, 67, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct action pathway in Arabidopsis. Plant Cell 2009, 21, 3152–3169. [Google Scholar] [CrossRef] [PubMed]
- Lomin, S.N.; Krivosheev, D.M.; Steklov, M.Y.; Arkhipov, D.V.; Osolodkin, D.I.; Schmülling, T.; Romanov, G.A. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J. Exp. Bot. 2015, 66, 1851–1863. [Google Scholar] [CrossRef] [PubMed]
- Spichal, L. Cytokinins—Recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012, 39, 267–284. [Google Scholar] [CrossRef]
- Jameson, P.E. Cytokinins. In Encyclopedia of Applied Plant Sciences; Thomas, B., Murray, B.G., Murphy, D.J., Eds.; Academic Press: Waltham, MA, USA, 2017; Volume 1, pp. 391–402. [Google Scholar]
- Nandi, S.K.; Palni, L.M.S.; Letham, D.S.; Knypl, J.S. The biosynthesis of cytokinins in germinating lupin seeds. J. Exp. Bot. 1988, 39, 1649–1665. [Google Scholar] [CrossRef]
- Nandi, S.K.; Palni, L.M.S. Transport and metabolism of dihydrozeatin riboside in germinating lupin seeds. J. Exp. Bot. 1989, 40, 615–629. [Google Scholar] [CrossRef]
- Hocart, C.H.; Letham, D.S. Biosynthesis of cytokinin in germinating seeds of Zea mays. J. Exp. Bot. 1990, 41, 1525–1528. [Google Scholar] [CrossRef]
- Nandi, S.K.; Palni, L.M.S.; de Klerk, G.J.M. The influence of the embryonic axis and cytokinins on reserve mobilization in germinating lupin seeds. J. Exp. Bot. 1995, 46, 329–336. [Google Scholar] [CrossRef]
- Villalobos, N.; Martin, L. Involvement of cytokinins in the germination of chick-pea seeds. Plant Growth Regul. 1992, 11, 277–291. [Google Scholar] [CrossRef]
- Munoz, J.L.; Martin, L.; Nicolas, G.; Villalobos, N. Influence of endogenous cytokinin on reserve mobilization in cotyledons of Cicer arietinum L. Plant Physiol. 1990, 93, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Green, T.R.; Baisted, D.J. Development of the activities of enzymes of the isoprenoid pathway during early stages of pea-seed germination. Biochem. J. 1972, 130, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Yadav, U.P.; Ayre, B.G.; Bush, D.R. Transgenic approaches to altering carbon and nitrogen partitioning in whole plants: Assessing the potential to improve crop yields and nutritional quality. Front. Plant Sci. 2015, 6, 275. [Google Scholar] [CrossRef] [PubMed]
- Petruzzelli, L.; Kunz, C.; Waldvogel, R.; Meins, F.; Leubner-Metzger, G. Distinct ethylene- and tissue-specific regulation of β-1,3-glucanases and chitinases during pea seed germination. Planta 1999, 209, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Nawa, Y.; Asahi, T. Relationship between the water content of pea cotyledons and mitochondrial development during the early stage of germination. Plant Cell Physiol. 1973, 14, 607–610. [Google Scholar]
- Barba-Espín, G.; Diaz-Vivancos, P.; Job, D.; Belghazi, M.; Job, C.; Hernández, A. Understanding the role of H2O2 during pea seed germination: A combined proteomic and hormone profiling approach. Plant Cell Environ. 2011, 34, 1907–1919. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D. Seed germination and dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Weitbrecht, K.; Müller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef] [PubMed]
- Hirose, N.; Takei, K.; Huroha, T.; Kamada-Nobusada, T.; Hayashi, H.; Sakakibara, H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 2008, 59, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Stirk, W.A.; Gold, J.D.; Novák, O.; Strnad, M.; van Staden, J. Changes in endogenous cytokinins during germination and seedling establishment of Tagetes minuta L. Plant Growth Regul. 2005, 47, 1–7. [Google Scholar] [CrossRef]
- Werner, T.; Holst, K.; Pörs, Y.; Guivarc'h, A.; Mustroph, A.; Chriqui, D.; Grimm, B.; Schmülling, T. Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J. Exp. Bot. 2008, 59, 2659–2672. [Google Scholar] [CrossRef] [PubMed]
- Ashikari, M.; Sakakibara, H.; Lin, S.Y.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Zalewski, W.; Gasparis, S.; Boczkowska, M.; Rajchel, I.; Orczyk, W.; Nadolska-Orczyk, A. Expression patterns of HvCKX genes indicate their role in growth and reproductive development of barley. PLoS ONE 2014. [Google Scholar] [CrossRef] [PubMed]
- Sanders, A.; Collier, R.; Trethewy, A.; Gould, G.; Sieker, R.; Tegeder, M. AAP1 regulates import of amino acids into developing Arabidopsis embryos. Plant J. 2009, 59, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Stransky, H.; Koch, W. The amino acid permease AAP8 is important for early seed development in Arabidopsis. Planta 2007, 226, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Klambt, D.; Thies, G.; Skoog, F. Isolation of cytokinins from Corynebacterium. fascians. Proc. Natl. Acad. Sci. USA 1966, 56, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Lawson, E.; Gantotti, B.; Starr, M. A 78-megadalton plasmid occurs in avirulent strains as well as virulent strains of Corynebacterium fascians. Curr. Microbiol. 1982, 7, 327–332. [Google Scholar] [CrossRef]
- Song, J.; Jiang, L.; Jameson, P.E. Co-ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. BMC Plant Biol. 2012, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Antoniadi, I.; Plačková, L.; Simonovik, B.; Doležal, K.; Turnbull, C.; Ljung, K.; Novák, O. Cell-type specific cytokinin distribution within the Arabidopsis primary root apex. Plant Cell 2015, 27, 1955–1967. [Google Scholar] [CrossRef] [PubMed]
| Cytokinin Levels | Cotyledon | Shoot | Root | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (pmol/g DW) | 4 hpi | 2 dpi | 5 dpi | 11 dpi | 15 dpi | 25 dpi | 5 dpi | 11 dpi | 15 dpi | 25 dpi | 5 dpi | 11 dpi | 15 dpi | 25 dpi |
| Total cytokinin | 5.05 ± 0.4 | 38.5 ± 7.3 | 51.37 ± 4.4 | 77.18 ± 11.4 | 47.23 ± 2.2 | 28.65 ± 2.2 | 357.41 ± 17.3 | 93.97 ± 14.9 | 55.83 ± 2.4 | 43.68 ± 1.3 | 197.89 ± 14.4 | 220.5 ± 26.8 | 113.68 ± 9.1 | 94.11 ± 5.9 |
| Total nucleotides | <lod | 34.5 ± 1.9 | 43.13 ± 3.8 | 64.73 ± 10.5 | 20.29 ± 2.8 | 9.39 ± 1.8 | 289.8 ± 13.6 | 65.44 ± 11.3 | 30.05 ± 2.8 | 17.64 ± 0.9 | 134.03 ± 13.3 | 162.86 ± 24.3 | 47.96 ± 5.0 | 31.61 ± 5.8 |
| Total bases | 3.72 ± 0.3 | 7.27 ± 0.6 | 3.72 ± 0.3 | 5.55 ± 0.5 | 2.75 ± 0.9 | 4.11 ± 0.3 | 17.31 ± 1.5 | 7.29 ± 1.1 | 5.9 ± 1.5 | 6.31 ± 0.5 | 12.65 ± 1.2 | 10.62 ± 0.3 | 8.97 ± 1.6 | 10.39 ± 0.5 |
| Total ribosides | 1.33 ± 0.1 | 3.56 ± 0.4 | 2.57 ± 0.4 | 3.07 ± 0.3 | 3.94 ± 0.4 | 2.36 ± 0.4 | 32.38 ± 3.3 | 8.36 ± 1.6 | 7.16 ± 0.5 | 3.71 ± 0.5 | 33.42 ± 2.0 | 26.24 ± 1.9 | 37 ± 8.2 | 13.87 ± 2.5 |
| Total O-glucosides | <lod | 2.08 ± 0.3 | 1.95 ± 0.2 | 3.75 ± 0.9 | 19.68 ± 0.9 | 11.81 ± 2.2 | 17.92 ± 1.5 | <lod | 12.49 ± 1.3 | 15.04 ± 1.8 | 17.48 ± 1.6 | 19.58 ± 1.3 | 19.03 ± 2.9 | 34.56 ± 4.2 |
| Total N-glucosides | <lod | <lod | <lod | 0.07 ± 0.02 | 0.64 ± 0.2 | 1.13 ± 0.4 | <lod | <lod | 0.24 ± 0.1 | 0.97 ± 0.1 | 0.34 ± 0.02 | 1.2 ± 0.3 | 0.73 ± 0.1 | 3.68 ± 0.4 |
| tZRMP | - | - | - | - | - | - | 23.87 ± 2.1 | 11.41 ± 1.8 | 2.19 ± 0.3 | 0.61 ± 0.04 | 15.17 ± 1.9 | 37.66 ± 6.9 | 6.68 ± 1.8 | 1.27 ± 0.4 |
| DHZRMP | <lod | <lod | <lod | 0.1 ± 0.01 | <lod | 0.27 ± 0.1 | 3.18 ± 0.1 | 0.66 ± 0.1 | 1.05 ± 0.3 | 0.89 ± 0.1 | 1.61 ± 0.3 | 0.83 ± 0.2 | 0.58 ± 0.1 | 0.26 ± 0.1 |
| iPRMP | - | - | - | - | - | - | 142.08 ± 9.9 | 22.66 ± 4.5 | 10.76 ± 1.3 | 6.91 ± 0.5 | 69 ± 4.02 | 68.53 ± 15.6 | 16.32 ± 1.0 | 10.09 ± 3.0 |
| cZRMP | <lod | 10.4 ± 1.2 | 17.25 ± 2.7 | 43 ± 9.1 | 15.07 ± 1.8 | 6.34 ± 1.1 | 120.66 ± 3.9 | 30.98 ± 6.6 | 17.76 ± 0.8 | 9.23 ± 0.5 | 48.25 ± 10.8 | 55.83 ± 9.0 | 24.46 ± 3.03 | 19.99 ± 3.1 |
| tZ | - | - | - | - | - | - | 7.16 ± 0.6 | 3.81 ± 0.8 | 1.74 ± 0.1 | 1.74 ± 0.1 | 4.55 ± 0.5 | 5.75 ± 0.5 | 3.83 ± 1.0 | 1.33 ± 0.1 |
| DHZ | 0.24 ± 0.04 | 0.1 ± 0.02 | 0.04 ± 0.01 | <lod | 0.09 ± 0.01 | 0.22 ± 0.1 | 0.49 ± 0.1 | <lod | 0.77 ± 0.1 | 1.26 ± 0.1 | 0.29 ± 0.1 | <lod | 0.41 ± 0.1 | 0.38 ± 0.02 |
| iP | - | - | - | - | - | - | 7.34 ± 0.7 | 2.79 ± 1.0 | 2.63 ± 1.0 | 2.41 ± 0.3 | 5.82 ± 0.8 | 3.62 ± 0.2 | 3.53 ± 0.6 | 3.22 ± 0.4 |
| cZ | 0.16 ± 0.03 | 0.15 ± 0.03 | 0.19 ± 0.02 | 1.01 ± 0.2 | 0.62 ± 0.1 | 0.68 ± 0.1 | 2.31 ± 0.3 | 0.69 ± 0.1 | 0.76 ± 0.1 | 0.91 ± 0.1 | 2.1 ± 0.2 | 1.25 ± 0.2 | 1.2 ± 0.1 | 5.46 ± 0.9 |
| tZR | - | - | - | - | - | - | 4.01 ± 0.3 | 1.86 ± 0.3 | 0.47 ± 0.1 | 0.28 ± 0.1 | 6.78 ± 0.4 | 9.13 ± 0.7 | 6.21 ± 1.4 | 0.75 ± 0.2 |
| DHZR | 0.17 ± 0.1 | 0.39 ± 0.1 | 0.15 ± 0.02 | 0.2 ± 0.03 | 0.12 ± 0.0 | 0.18 ± 0.04 | 2.85 ± 0.3 | 1.65 ± 0.4 | 0.69 ± 0.1 | 0.63 ± 0.1 | 2.18 ± 0.1 | 2.28 ± 0.2 | 0.85 ± 0.2 | 0.8 ± 0.2 |
| iPR | - | - | - | - | - | - | 13.12 ± 2.5 | 2.88 ± 0.8 | 2.98 ± 0.8 | 1.33 ± 0.1 | 19.19 ± 1.7 | 11.24 ± 1.9 | 24.44 ± 5.8 | 7.81 ± 2.0 |
| cZR | 0.89 ± 0.1 | 0.65 ± 0.03 | 0.77 ± 0.1 | 1.57 ± 0.2 | 2.66 ± 0.5 | 1.19 ± 0.3 | 12.41 ± 2.3 | 1.97 ± 0.3 | 3.02 ± 0.8 | 1.47 ± 0.3 | 5.27 ± 0.1 | 3.59 ± 0.5 | 5.5 ± 1.0 | 4.51 ± 0.3 |
| tZOG | - | - | - | - | - | - | 1.42 ± 0.14 | 4.03 ± 0.8 | 1.74 ± 0.14 | 1.5 ± 0.1 | 0.73 ± 0.1 | 4.04 ± 0.6 | 1.94 ± 0.2 | 0.9 ± 0.1 |
| DHZOG | <lod | <lod | 0.03 ± 0.0 | 0.15 ± 0.02 | 0.3 ± 0.03 | 0.31 ± 0.04 | 0.32 ± 0.04 | 1.25 ± 0.2 | 2.71 ± 0.3 | 6.79 ± 1.1 | 0.19 ± 0.01 | 1.41 ± 0.1 | 1.05 ± 0.1 | 0.34 ± 0.03 |
| cZOG | <lod | 1.26 ± 0.1 | 0.98 ± 0.1 | 1.76 ± 0.4 | 9.63 ± 0.3 | 4.45 ± 0.8 | 2.54 ± 0.4 | 2.52 ± 0.4 | 2.93 ± 0.2 | 3.15 ± 0.3 | 2.83 ± 0.2 | 3.54 ± 0.6 | 2.24 ± 0.2 | 3.77 ± 0.5 |
| tZROG | - | - | - | - | - | - | 1.35 ± 0.1 | 1.25 ± 0.2 | 1.05 ± 0.1 | 1.03 ± 0.1 | 0.7 ± 0.03 | 1.12 ± 0.2 | 1.3 ± 0.32 | 0.7 ± 0.1 |
| DHZROG | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod |
| cZROG | <lod | 0.74 ± 0.2 | 0.66 ± 0.1 | 1.29 ± 0.4 | 8.39 ± 0.7 | 6.22 ± 1.6 | 12.3 ± 1.5 | 3.84 ± 0.5 | 4.05 ± 0.8 | 2.58 ± 0.3 | 13.04 ± 1.4 | 9.48 ± 0.8 | 12.5 ± 2.9 | 28.85 ± 3.9 |
| tZ9G | <lod | <lod | <lod | 0.12 ± 0.02 | 0.64 ± 0.2 | 1.13 ± 0.4 | <lod | <lod | 0.24 ± 0.1 | 0.97 ± 0.1 | <lod | <lod | 0.39 ± 0.1 | 2.88 ± 0.3 |
| DHZ9G | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod |
| iP9G | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod |
| cZ9G | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | <lod | 0.34 ± 0.02 | 0.24 ± 0.01 | 0.34 ± 0.1 | 0.81 ± 0.1 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jameson, P.E.; Dhandapani, P.; Novak, O.; Song, J. Cytokinins and Expression of SWEET, SUT, CWINV and AAP Genes Increase as Pea Seeds Germinate. Int. J. Mol. Sci. 2016, 17, 2013. https://doi.org/10.3390/ijms17122013
Jameson PE, Dhandapani P, Novak O, Song J. Cytokinins and Expression of SWEET, SUT, CWINV and AAP Genes Increase as Pea Seeds Germinate. International Journal of Molecular Sciences. 2016; 17(12):2013. https://doi.org/10.3390/ijms17122013
Chicago/Turabian StyleJameson, Paula E., Pragatheswari Dhandapani, Ondrej Novak, and Jiancheng Song. 2016. "Cytokinins and Expression of SWEET, SUT, CWINV and AAP Genes Increase as Pea Seeds Germinate" International Journal of Molecular Sciences 17, no. 12: 2013. https://doi.org/10.3390/ijms17122013







