Validating Intravascular Imaging with Serial Optical Coherence Tomography and Confocal Fluorescence Microscopy
Abstract
:1. Introduction
2. Results
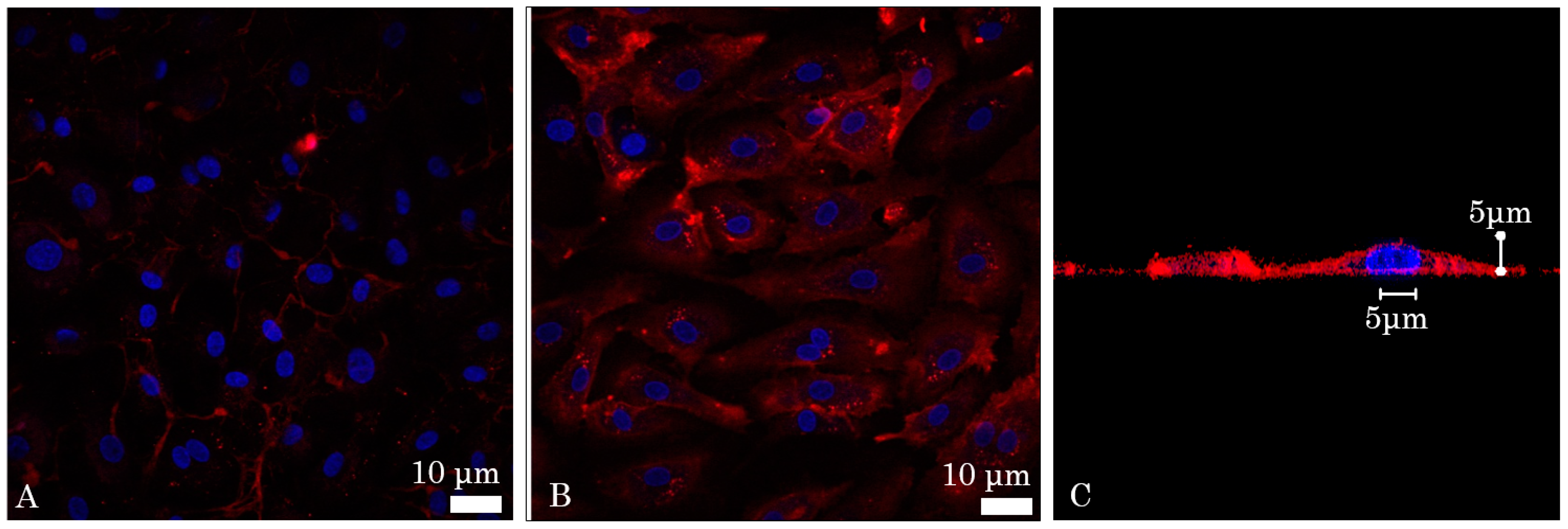
2.1. In Vitro Affinity of Anti-ICAM-1 Antibody
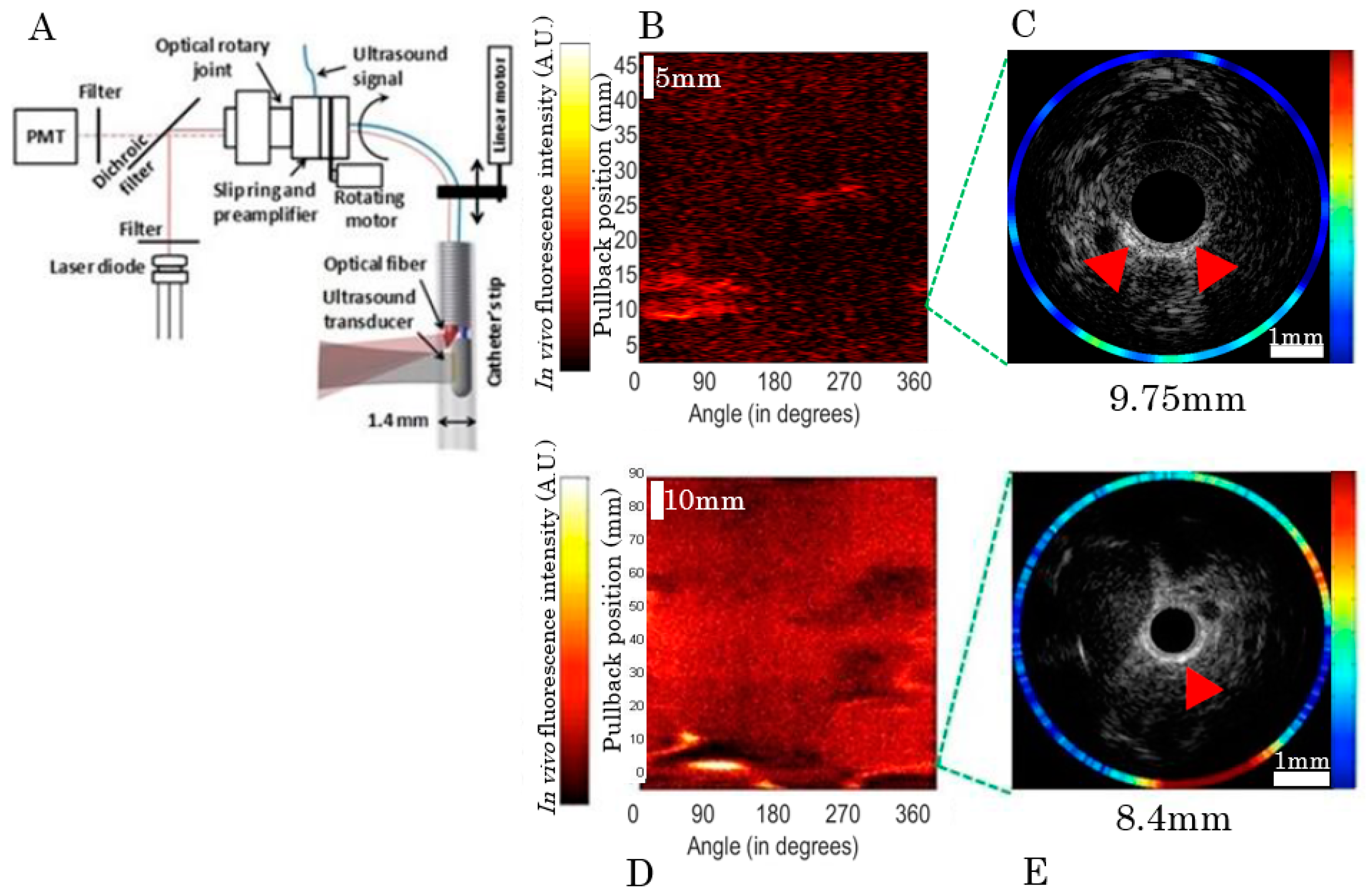
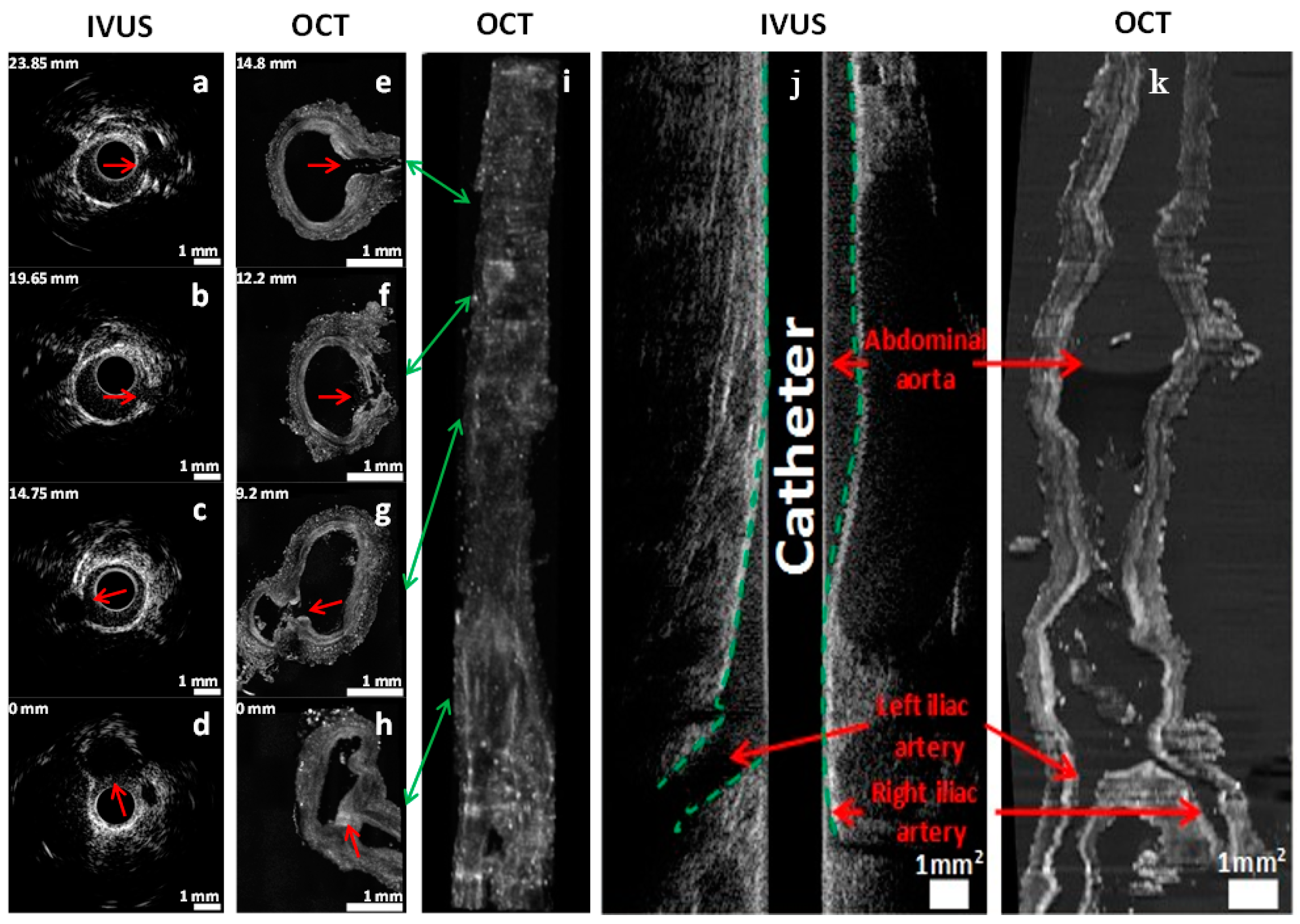
2.2. In Vivo Catheter Imaging
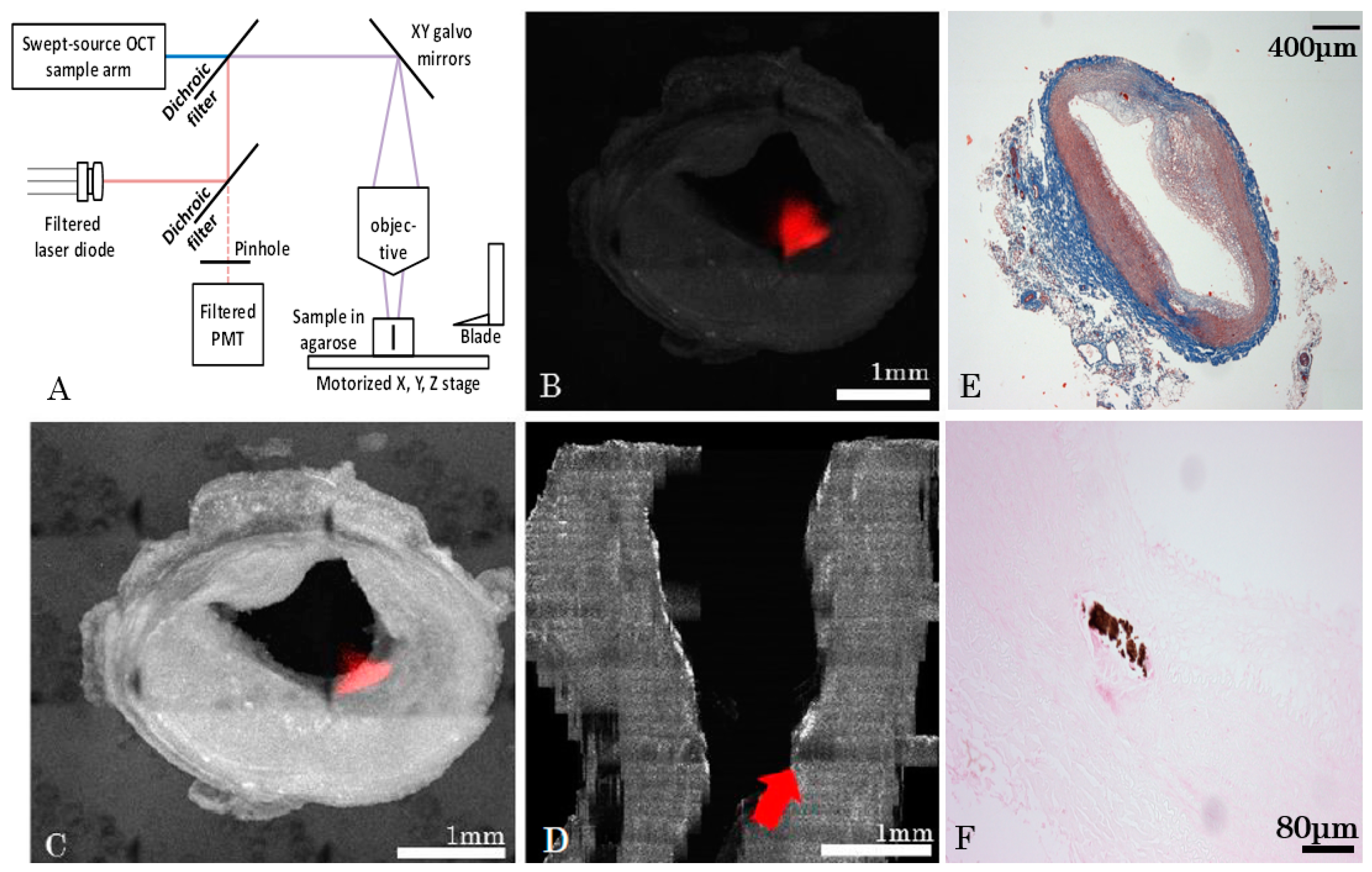
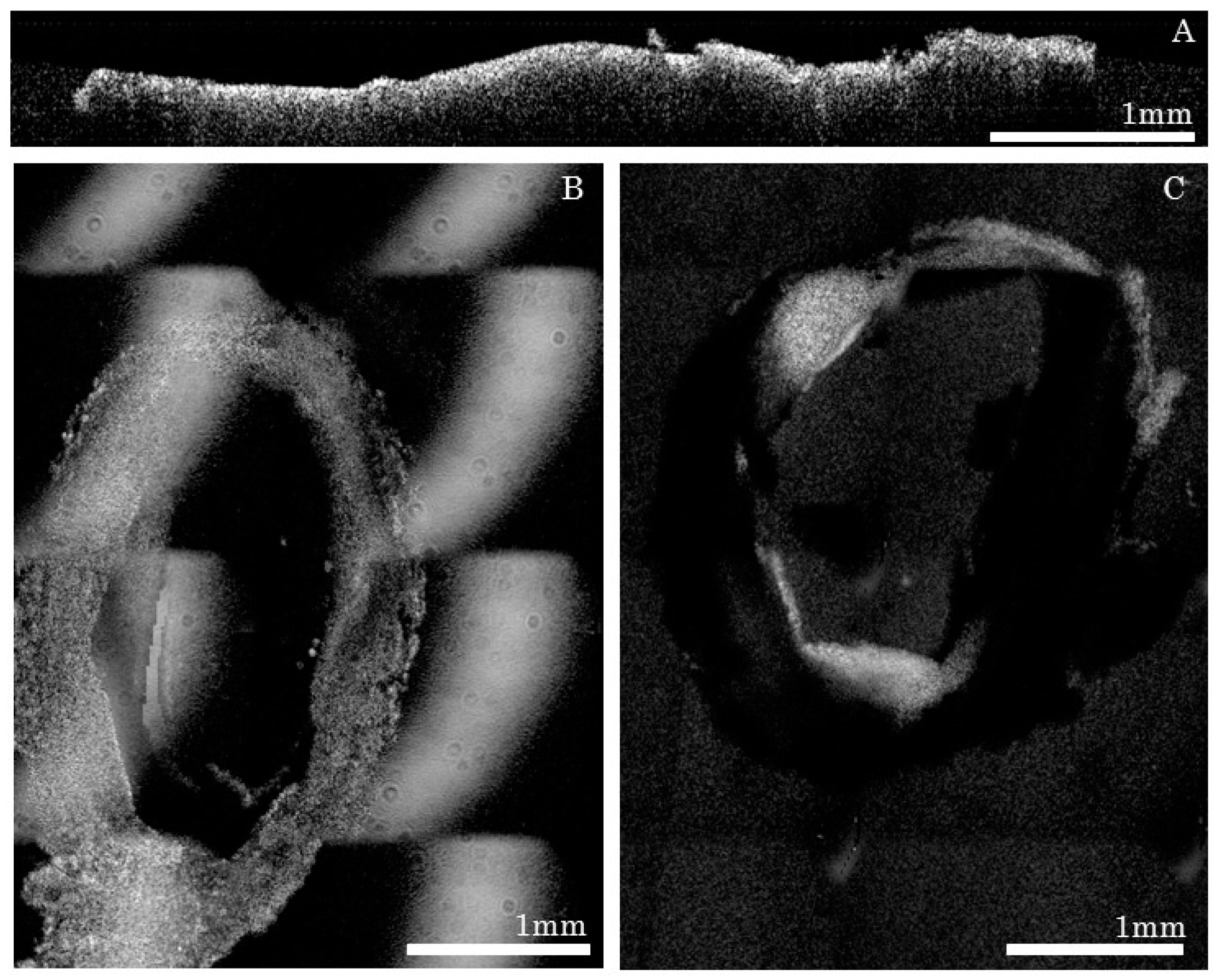
2.3. Serial Imaging System, Ex Vivo OCT Reconstructions and Fluorescence Alignment
Data Reconstruction
- For each tissue slice, the volume position in the mosaic reference frame was estimated using microscope acquisition data;
- Volumes were then stitched for each tissue slice;
- Post processing was performed that included cropping of the field of view, identification of lumen mask and Beer-Lambert intensity correction;
- After post processing steps, slices were assembled together in order to form a longitudinal 3D volume.
2.4. Alignment and Tissue Deformation
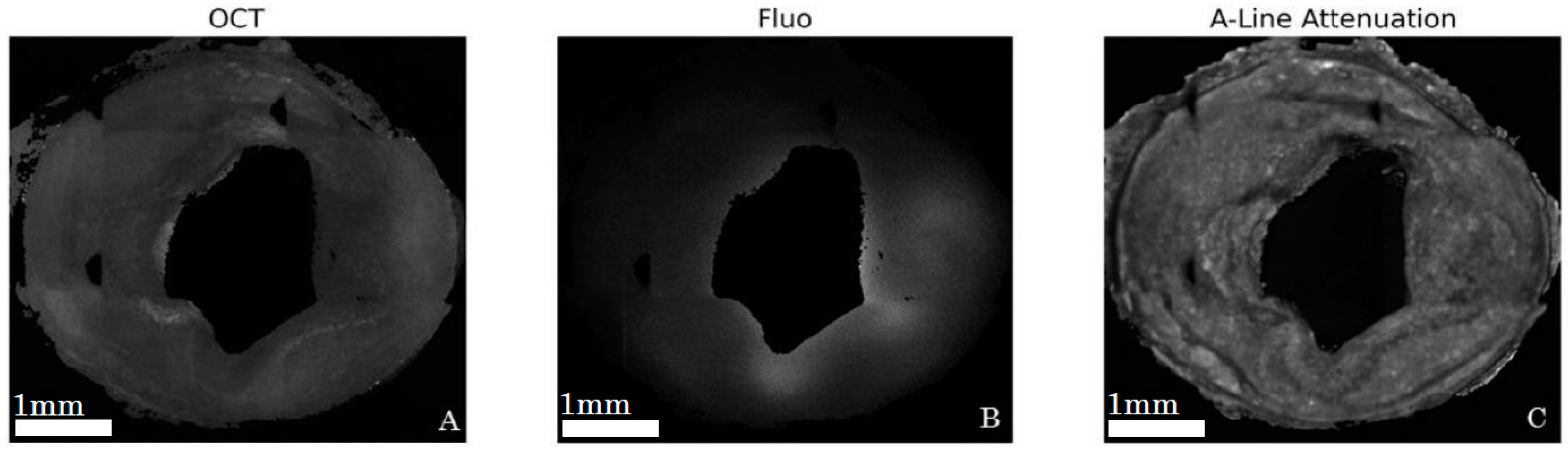
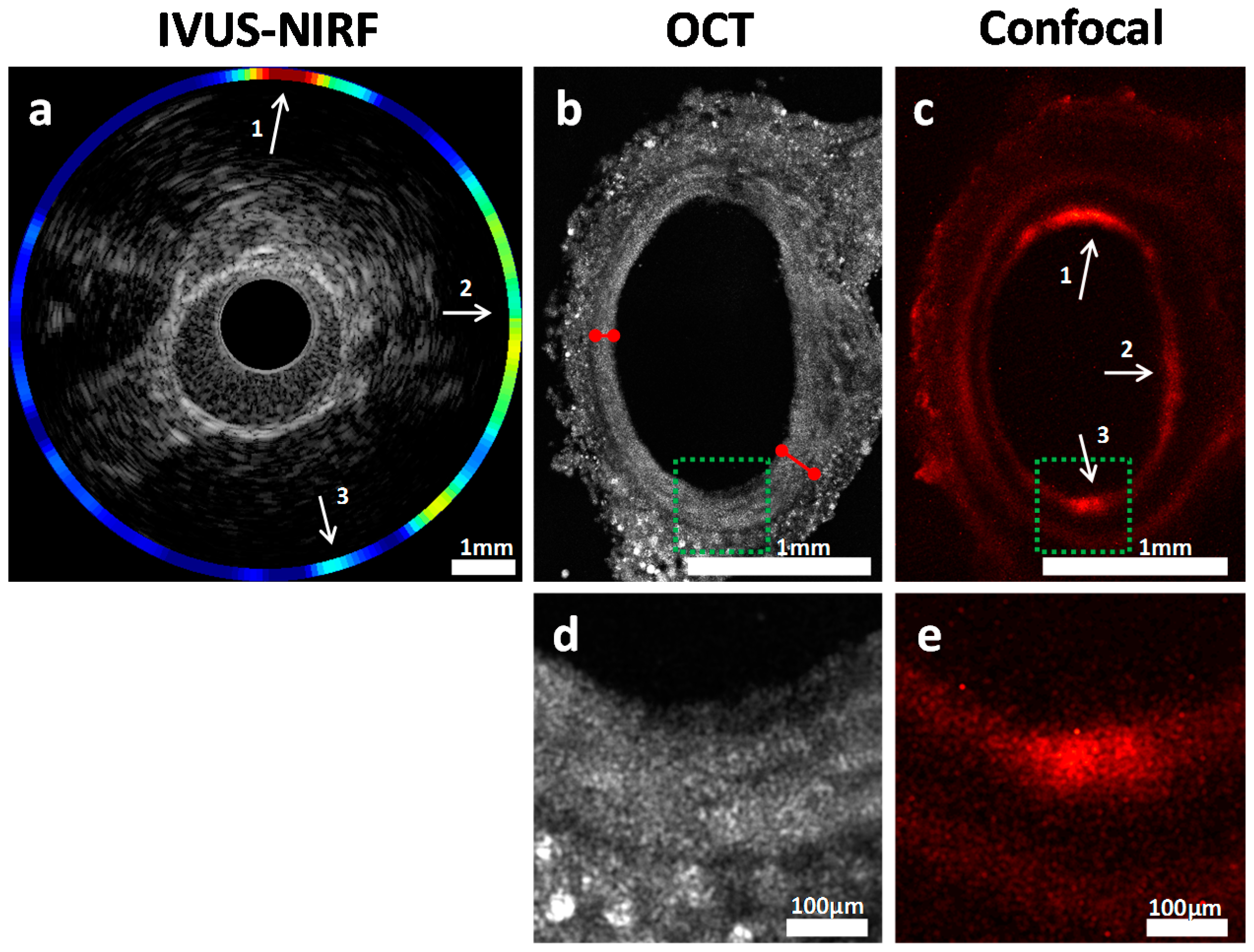
2.5. Validation of Intravascular Molecular Imaging
3. Discussion
3.1. Slicing Optimization
3.2. Optical Optimization
3.3. Big-Data Processing
4. Materials and Methods
4.1. Animal Model for Atherosclerosis
4.2. Balloon Dilatation Procedures
4.3. Bimodal Near-Infrared Fluorescence (NIRF)/Intravascular Ultrasound (IVUS) Imaging Catheter System
4.4. In Vivo NIRF Imaging Procedure
4.5. Ex Vivo Imaging System and Methods
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Libby, P.; Okamoto, Y.; Rocha, V.Z.; Folco, E. Inflammation in atherosclerosis transition from theory to practice. Circ. J. 2010, 74, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Ridker, P.M.; Hansson, G.K. Progress and challenges in translating the biology of atherosclerosis. Nature 2011, 473, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Bentzon, J.F.; Otsuka, F.; Virmani, R.; Falk, E. Mechanisms of plaque formation and rupture. Circ. Res. 2014, 114, 1852–1866. [Google Scholar] [CrossRef] [PubMed]
- Muller, J.E.; Tofler, G.H.; Stone, P.H. Circadian variation and triggers of onset of acute cardiovascular disease. Circulation 1989, 79, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Mintz, G.S.; Painter, J.A.; Pichard, A.D.; Kent, K.M.; Satler, L.F.; Popma, J.J.; Chuang, Y.C.; Bucher, T.A.; Sokolowicz, L.E.; Leon, M.B. Atherosclerosis in angiographically “normal” coronary artery reference segments: An intravascular ultrasound study with clinical correlations. J. Am. Coll. Cardiol. 1995, 25, 1479–1485. [Google Scholar] [CrossRef]
- Tardif, J.C.; Lesage, F.; Harel, F.; Romeo, P.; Pressacco, J. Imaging biomarkers in atherosclerosis trials. Circ. Cardiovasc. Imaging 2011, 4, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Berry, C.L.; L’Allier, P.; Grégoire, J.; Lespérance, J.; Lévesque, S.; Ibrahim, R.; Tardif, J.C. Comparison of intravascular ultrasound and quantitative coronary angiography for the assessment of coronary artery disease progression. Circulation 2007, 115, 1851–1857. [Google Scholar] [CrossRef] [PubMed]
- Mintz, G.S.; Garcia-Garcia, H.M.; Nicholls, S.J.; Weissman, N.J.; Bruining, N.; Crowe, T.; Tardif, J.C.; Serruys, P.W. Clinical experts consensus document on standards of acquisition, measurement and reporting of intravascular ultrasound regression/progression studies. EuroIntervention 2011, 6, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, O.D.; Sakakura, K.; Otsuka, F.; Yahagi, K.; Virmani, R.; Joner, M. Expectations and limitations in contemporary intravascular imaging: Lessons learned from pathology. Expert Rev Cardiovasc. Ther. 2014, 12, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Abran, M.; Cloutier, G.; Cardinal, M.H.R.; Chayer, B.; Tardif, J.C.; Lesage, F. Development of a photoacoustic ultrasound and fluorescence imaging catheter for the study of atherosclerotic plaque. IEEE Trans. Biomed. Circuits Syst. 2014, 8, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Dixon, A.J.; Hossack, J. Intravascular near-infrared fluorescence catheter with ultrasound guidance and blood attenuation correction. J. Biomed. Opt. 2013, 18. [Google Scholar] [CrossRef] [PubMed]
- Mallas, G.; Brooks, D.H.; Rosenthal, A.; Nudelman, R.N.; Mauskapf, A.; Jaffer, F.A.; Ntziachristos, V. Improving quantification of intravascular fluorescence imaging using structural information. Phys. Med. Biol. 2012, 57, 6395–6406. [Google Scholar] [CrossRef] [PubMed]
- Bec, J.; Xie, H.; Yankelevich, D.R.; Zhou, F.; Sun, Y.; Ghata, N.; Aldredge, R.; Marcu, L. Design construction and validation of a rotary multifunctional intravascular diagnostic catheter combining multispectral fluorescence lifetime imaging and intravascular ultrasound. J. Biomed. Opt. 2011, 4, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Jaffer, F.A.; Calfon, M.A.; Rosenthal, A.; Mallas, G.; Razansky, R.N.; Mauskapf, A.; Weissleder, R.; Libby, P.; Ntziachristos, V. Two-dimensional intravascular near-infrared fluorescence molecular imaging of inflammation in atherosclerosis and stent-induced vascular injury. J. Am. Coll. Cardiol. 2011, 57, 2516–2526. [Google Scholar] [CrossRef] [PubMed]
- Jaffer, F.A.; Vinegoni, C.; John, M.C.; Aikawa, E.; Gold, H.K.; Finn, A.V.; Ntziachristos, V.; Libby, P.; Weissleder, R. Real-time catheter molecular sensing of inflammation in proteolytically active atherosclerosis. Circulation 2008, 118, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Nahrendorf, M.; Jaffer, F.A.; Kelly, K.A.; Sosnovik, D.E.; Aikawa, E.; Libby, P.; Weissleder, R. Noninvasive vascular cell adhesion molecule-1 imaging identifies inflammatory activation of cells in atherosclerosis. Circulation 2006, 114, 1504–1511. [Google Scholar] [CrossRef] [PubMed]
- Rouleau, L.; Berti, R.; Ng, V.W.K.; Matteau-Pelletier, C.; Lam, T.; Saboural, P.; Kakkar, A.K.; Lesage, F.; Rhéaume, E.; Tardif, J.C. VCAM-1-targeting gold nanoshell probe for photoacoustic imaging of atherosclerotic plaque in mice. Contrast Med. Mol. Imaging 2013, 8, 27–39. [Google Scholar] [CrossRef] [PubMed]
- De Vries, B.M.W.; Hillebrands, J.; van Dam, G.M.; Tio, R.A.; de Jong, J.S.; Slart, R.H.J.A.; Zeebregts, C.J. Images in cardiovascular medicine. Multispectral near-infrared fluorescence molecular imaging of matrix metalloproteinases in a human carotid plaque using a matrix-degrading metalloproteinase-sensitive activatable fluorescent probe. Circulation 2009, 119, e534–e536. [Google Scholar] [CrossRef] [PubMed]
- Abran, M.; Stähli, B.E.; Merlet, N.; Mihalache-Avram, T.; Mecteau, M.; Rhéaume, E.; Busseuil, D.; Tardif, J.C.; Lesage, F. Validating a bimodal intravascular ultrasound (IVUS) and near-infrared fluorescence (NIRF) catheter for atherosclerotic plaque detection in rabbits. Biomed. Opt. Express 2015, 6, 3989–3999. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhu, J.; Akkin, T. Serial optical coherence scanner for large-scale brain imaging at microscopic resolution. NeuroImage 2014, 84, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Castonguay, A.; Avti, P.K.; Moeini, M.; Pouliot, P.; Tabatabaei, M.S.; Bélanger, S.; Lesage, F. Investigating the correlation between white matter and microvasculature in aging using large scale optical coherence tomography and confocal fluorescence imaging combined with tissue sectioning. Proc. SPIE 2015, 9328. [Google Scholar] [CrossRef]
- Pande, A.N.; Kohler, R.H.; Aikawa, E.; Weissleder, R.; Jaffer, F.A. Detection of macrophage activity in atherosclerosis in vivo using multichannel, high-resolution laser scanning fluorescence microscopy. J. Biomed. Opt. 2006, 11. [Google Scholar] [CrossRef] [PubMed]
- Preibisch, S.; Saalfeld, S. Fast stitching of large 3d biological datasets. In Proceedings of the 2nd ImageJ User and Developer Conference, Luxembourg, 7–8 November 2008.
- Faber, D.; van der Meer, F.; Aalders, M.; van Leeuwen, T. Quantitative measurement of attenuation coefficients of weakly scattering media using optical coherence tomography. Opt. Express 2004, 12, 4353–4365. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tardif, P.-L.; Bertrand, M.-J.; Abran, M.; Castonguay, A.; Lefebvre, J.; Stähli, B.E.; Merlet, N.; Mihalache-Avram, T.; Geoffroy, P.; Mecteau, M.; et al. Validating Intravascular Imaging with Serial Optical Coherence Tomography and Confocal Fluorescence Microscopy. Int. J. Mol. Sci. 2016, 17, 2110. https://doi.org/10.3390/ijms17122110
Tardif P-L, Bertrand M-J, Abran M, Castonguay A, Lefebvre J, Stähli BE, Merlet N, Mihalache-Avram T, Geoffroy P, Mecteau M, et al. Validating Intravascular Imaging with Serial Optical Coherence Tomography and Confocal Fluorescence Microscopy. International Journal of Molecular Sciences. 2016; 17(12):2110. https://doi.org/10.3390/ijms17122110
Chicago/Turabian StyleTardif, Pier-Luc, Marie-Jeanne Bertrand, Maxime Abran, Alexandre Castonguay, Joël Lefebvre, Barbara E. Stähli, Nolwenn Merlet, Teodora Mihalache-Avram, Pascale Geoffroy, Mélanie Mecteau, and et al. 2016. "Validating Intravascular Imaging with Serial Optical Coherence Tomography and Confocal Fluorescence Microscopy" International Journal of Molecular Sciences 17, no. 12: 2110. https://doi.org/10.3390/ijms17122110




