Modulation of GLO1 Expression Affects Malignant Properties of Cells
Abstract
:1. Introduction
2. Results
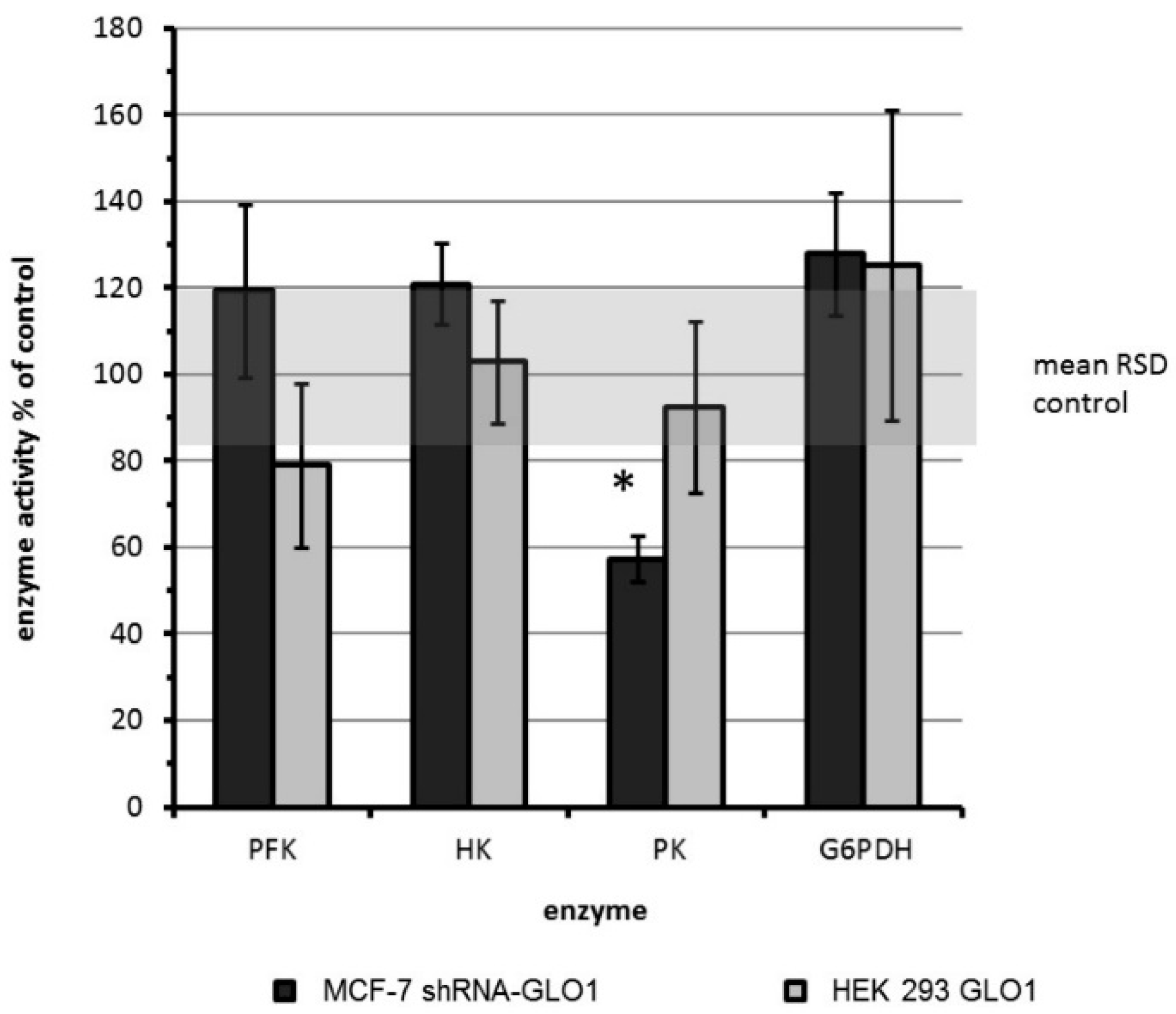
2.1. Modulation of GLO1 Enzyme Activity Impacts Downstream Glycolytic Enzymes
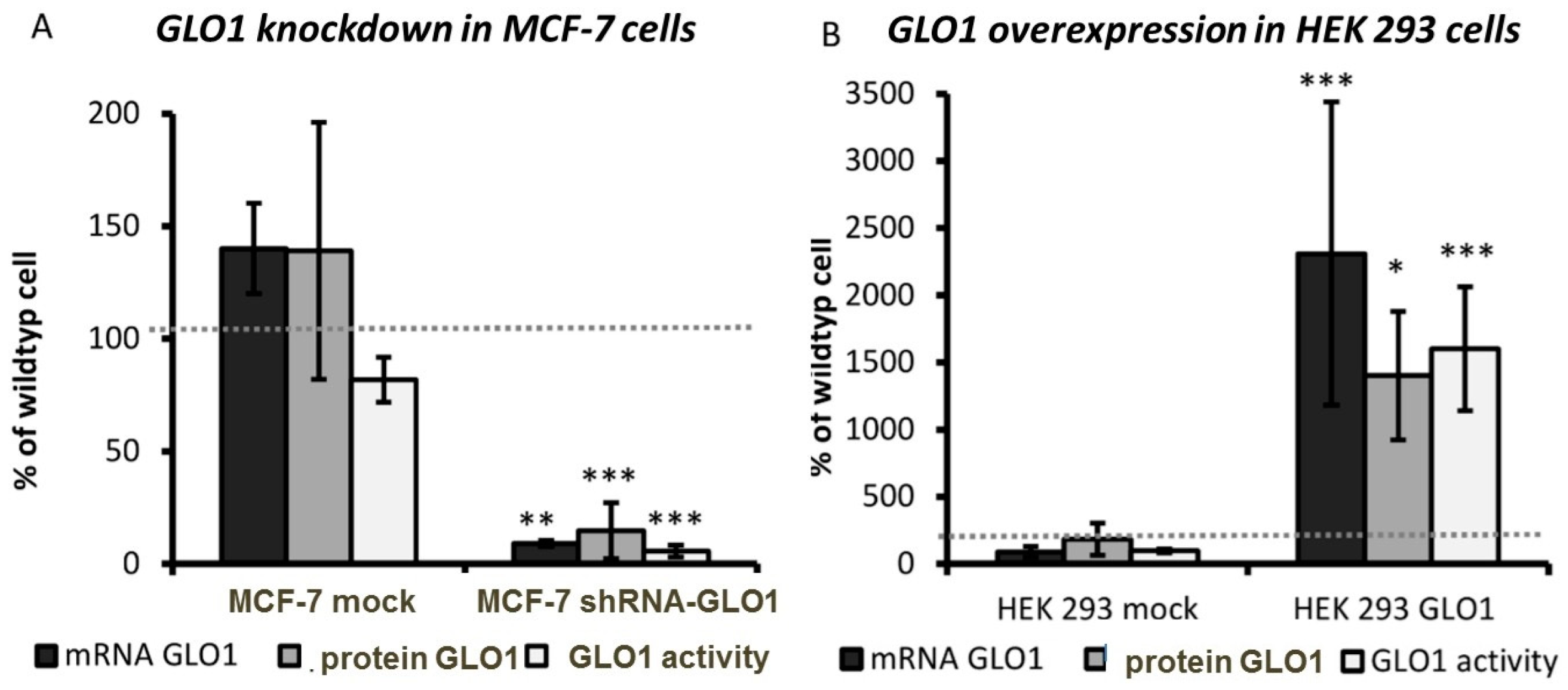
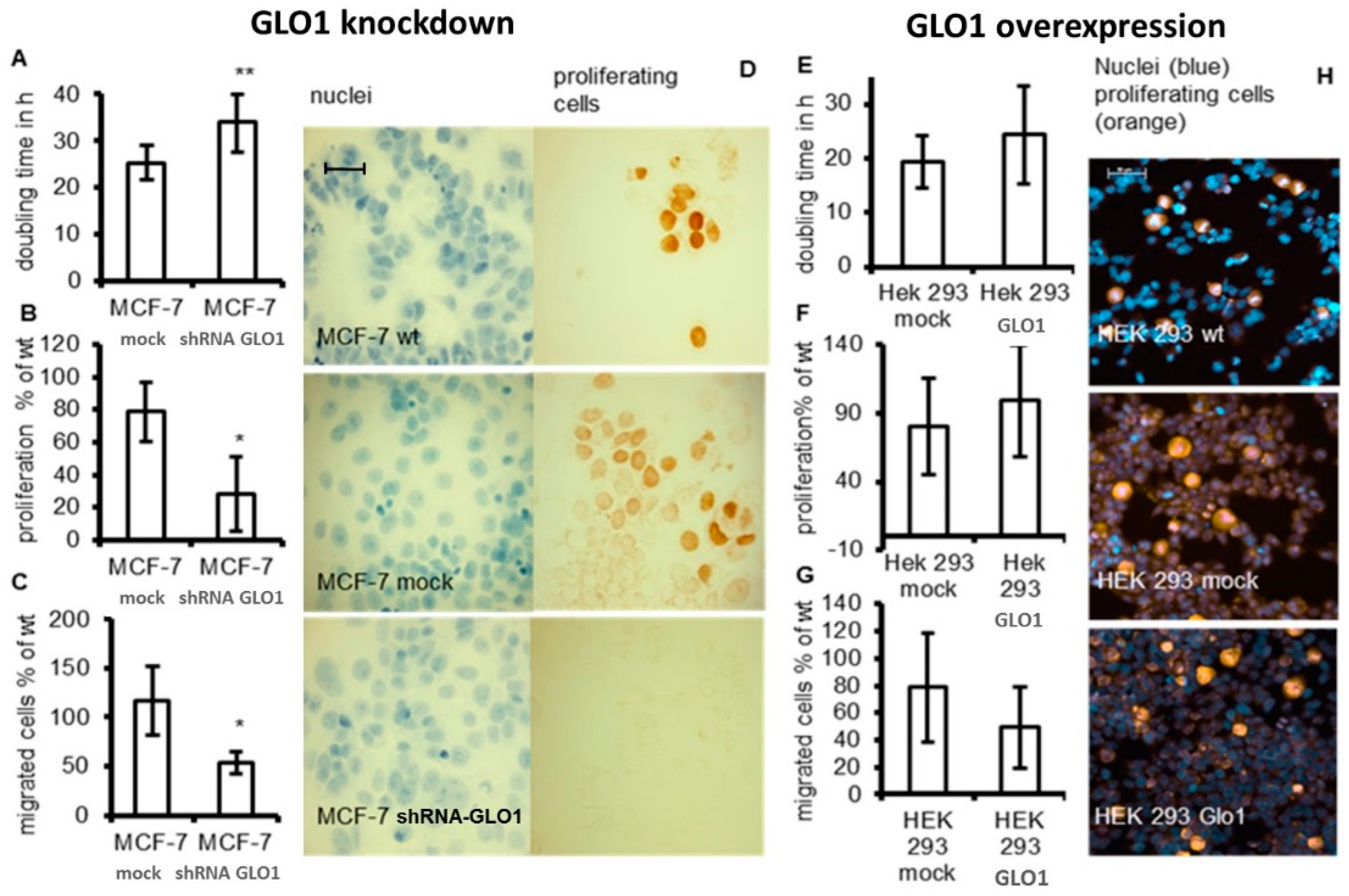
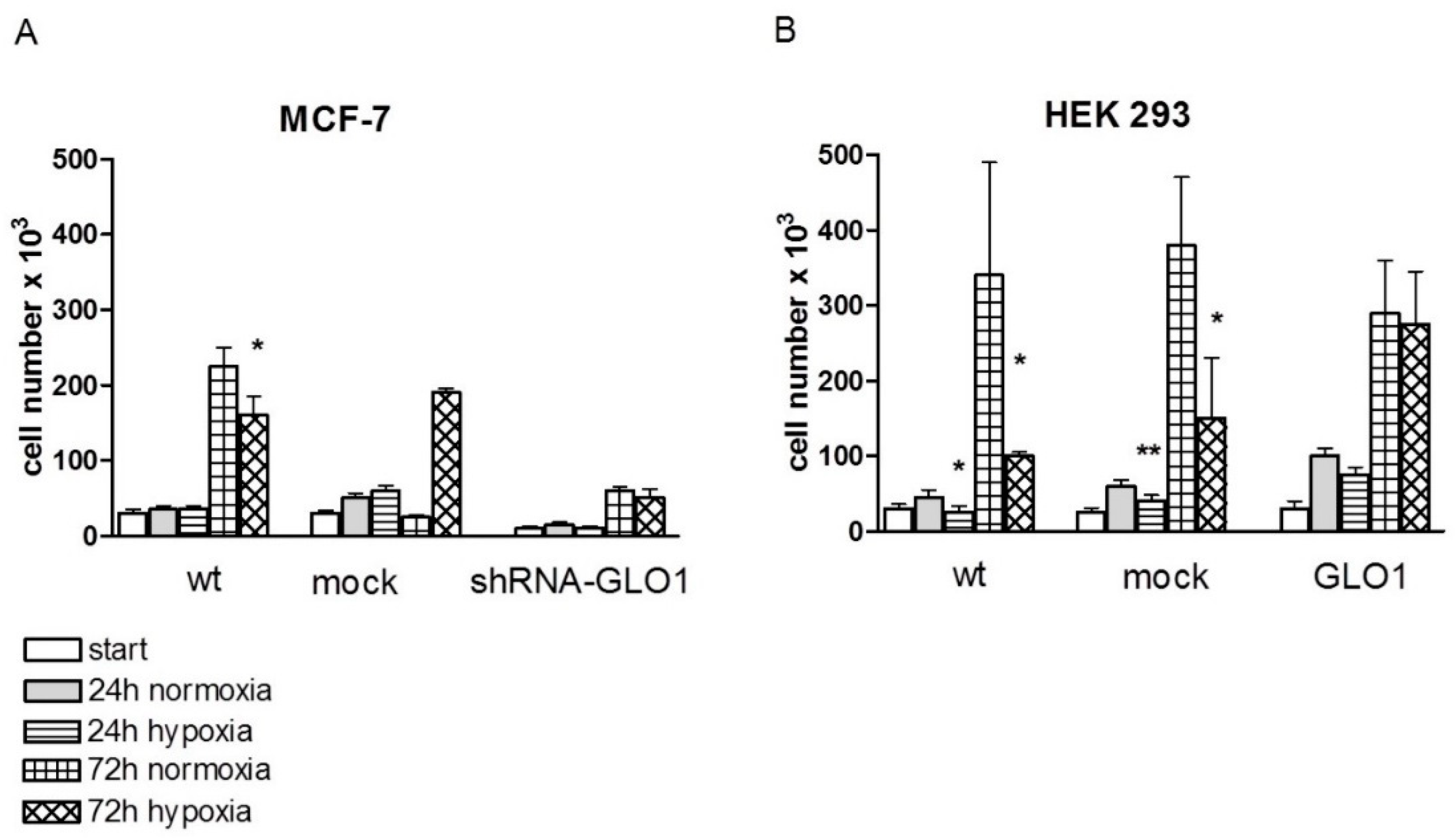
2.2. Tumor-Related Physiological Parameters Are Affected by GLO1-Knockdown But Not by GLO1-Overexpression
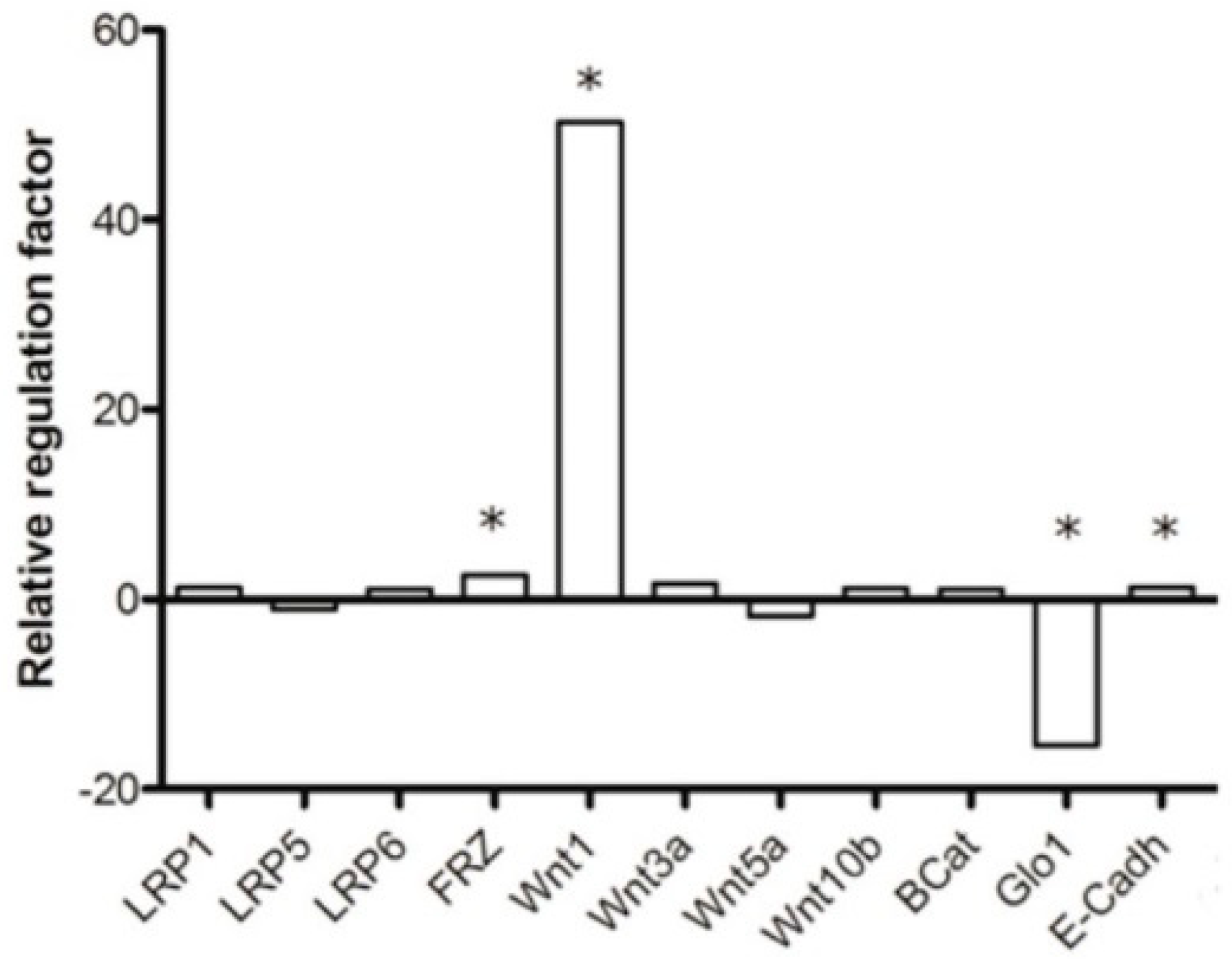
2.3. GLO1-Overexpressing Cells Show Better Adaption to Hypoxia
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture
4.3. Protein Extraction and Determination of Protein Content
4.4. Construction of shRNA Expressing Plasmids for GLO1 Silencing
4.5. Cloning of GLO1-Overexpressing Plasmids
4.6. Generation of Stably Transfected Cell Lines
4.7. Total RNA Isolation and RT-PCR
4.8. Western Blot Analysis
4.9. Immunohistochemistry
4.10. Doubling Time, Proliferation and Vitality Assay
4.11. Determination of ATP in Cell Lysates and NADPH Production
4.12. Migration Assay
4.13. Anchorage-Independent Growth
4.14. Enzyme Activity Measurements
4.15. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McLellan, A.C.; Thornalley, P.J. Glyoxalase activity in human red blood cells fractioned by age. Mech. Ageing Dev. 1989, 48, 63–71. [Google Scholar] [CrossRef]
- Thornalley, P.J. Glyoxalase I—Structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 2003, 31, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J.; Waris, S.; Fleming, T.; Santarius, T.; Larkin, S.J.; Winklhofer-Roob, B.M.; Stratton, M.R.; Rabbani, N. Imidazopurinones are markers of physiological genomic damage linked to DNA instability and glyoxalase 1-associated tumour multidrug resistance. Nucleic Acids Res. 2010, 38, 5432–5442. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J.; Rabbani, N. Glyoxalase in tumourigenesis and multidrug resistance. Semin. Cell Dev. Biol. 2011, 22, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Neubert, D.; Peters, H.; Teske, S.; Kohler, E.; Barrach, H.J. Studis on the problem of “aerobic glycolysis” occuring in mammalian embryos. Naunyn-Schmiedeberg’s Arch. Pharmacol. 1971, 268, 235–241. [Google Scholar] [CrossRef]
- Dang, C.V.; Semenza, G.L. Oncogenic alterations of metabolism. Trends Biochem. Sci. 1999, 24, 68–72. [Google Scholar] [CrossRef]
- Cuezva, J.M.; Krajewska, M.; de Heredia, M.L.; Krajewski, S.; Santamaria, G.; Kim, H.; Zapata, J.M.; Marusawa, H.; Chamorro, M.; Reed, J.C. The bioenergetic signature of cancer: A marker of tumor progression. Cancer Res. 2002, 62, 6674–6681. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, S.; Tew, K.D. Analysis of glyoxalase-I from normal and tumor tissue from human colon. Biochim. Biophys. Acta 1993, 1182, 311–316. [Google Scholar] [CrossRef]
- Rulli, A.; Carli, L.; Romani, R.; Baroni, T.; Giovannini, E.; Rosi, G.; Talesa, V. Expression of glyoxalase I and ii in normal and breast cancer tissues. Breast Cancer Res. Treat. 2001, 66, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Davidson, S.D.; Cherry, J.P.; Choudhury, M.S.; Tazaki, H.; Mallouh, C.; Konno, S. Glyoxalase I activity in human prostate cancer: A potential marker and importance in chemotherapy. J. Urol. 1999, 161, 690–691. [Google Scholar] [CrossRef]
- Bair, W.B., 3rd; Cabello, C.M.; Uchida, K.; Bause, A.S.; Wondrak, G.T. GLO1 overexpression in human malignant melanoma. Melanoma Res. 2010, 20, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, F.; Zaman, M.; Thornalley, P.; Masters, J. Glyoxalase activities in human tumour cell lines in vitro. Anticancer Res. 1993, 13, 151–155. [Google Scholar] [PubMed]
- Santarius, T.; Bignell, G.R.; Greenman, C.D.; Widaa, S.; Chen, L.; Mahoney, C.L.; Butler, A.; Edkins, S.; Waris, S.; Thornalley, P.J.; et al. GLO1—A novel amplified gene in human cancer. Genes Chromosomes Cancer 2010, 49, 711–725. [Google Scholar] [CrossRef] [PubMed]
- Fonseca-Sánchez, M.A.; Rodríguez Cuevas, S.; Mendoza-Hernández, G.; Bautista-Piña, V.; Arechaga Ocampo, E.; Hidalgo Miranda, A.; Quintanar Jurado, V.; Marchat, L.A.; Álvarez-Sánchez, E.; Pérez Plasencia, C.; et al. Breast cancer proteomics reveals a positive correlation between glyoxalase 1 expression and high tumor grade. Int. J. Oncol. 2012, 41, 670–680. [Google Scholar] [PubMed]
- Ranganathan, S.; Walsh, E.S.; Tew, K.D. Glyoxalase I in detoxification: Studies using a glyoxalase I transfectant cell line. Biochem. J. 1995, 309, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Davies, G.F.; Roesler, W.J.; Juurlink, B.H.; Harkness, T.A. Troglitazone overcomes doxorubicin-resistance in resistant k562 leukemia cells. Leuk. Lymphoma 2005, 46, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Sharkey, E.M.; O′Neill, H.B.; Kavarana, M.J.; Wang, H.; Creighton, D.J.; Sentz, D.L.; Eiseman, J.L. Pharmacokinetics and antitumor properties in tumor-bearing mice of an enediol analogue inhibitor of glyoxalase I. Cancer Chemother. Pharmacol. 2000, 46, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, H.; Mashima, T.; Sato, S.; Hashimoto, Y.; Yamori, T.; Tsuruo, T. Selective activation of apoptosis program by S-p-bromobenzylglutathione cyclopentyl diester in glyoxalase I-overexpressing human lung cancer cells. Clin. Cancer Res. 2001, 7, 2513–2518. [Google Scholar] [PubMed]
- Santel, T.; Pflug, G.; Hemdan, N.Y.; Schafer, A.; Hollenbach, M.; Buchold, M.; Hintersdorf, A.; Lindner, I.; Otto, A.; Bigl, M.; et al. Curcumin inhibits glyoxalase 1: A possible link to its anti-inflammatory and anti-tumor activity. PLoS ONE 2008, 3, e3508. [Google Scholar] [CrossRef] [PubMed]
- Mulukutla, B.C.; Khan, S.; Lange, A.; Hu, W.S. Glucose metabolism in mammalian cell culture: New insights for tweaking vintage pathways. Trends Biotechnol. 2010, 28, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.N.; Braunstein, L.D.; Pang, J.; Stuhlmeier, K.M.; Xi, Q.C.; Tian, X.; Stanton, R.C. Importance of glucose-6-phosphate dehydrogenase activity for cell growth. J. Biol. Chem. 1998, 273, 10609–10617. [Google Scholar] [CrossRef] [PubMed]
- Kalapos, M.P. Methylglyoxal in living organisms: Chemistry, biochemistry, toxicology and biological implications. Toxicol. Lett. 1999, 110, 145–175. [Google Scholar] [CrossRef]
- MacDonald, B.T.; Tamai, K.; He, X. WNT/β-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, T.; Nangaku, M.; Kojima, I.; Nagai, R.; Ingelfinger, J.R.; Miyata, T.; Fujita, T.; Inagi, R. Glyoxalase I overexpression ameliorates renal ischemia-reperfusion injury in rats. Am. J. Physiol. Renal Physiol. 2009, 296, F912–F921. [Google Scholar] [CrossRef] [PubMed]
- Fantin, V.R.; St-Pierre, J.; Leder, P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 2006, 9, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Edwards, L.G.; Thornalley, P.J. Effect of methylglyoxal on human leukaemia 60 cell growth: Modification of DNA G1 growth arrest and induction of apoptosis. Leuk. Res. 1996, 20, 397–405. [Google Scholar] [CrossRef]
- Biswas, S.; Ray, M.; Misra, S.; Dutta, D.P.; Ray, S. Selective inhibition of mitochondrial respiration and glycolysis in human leukaemic leucocytes by methylglyoxal. Biochem. J. 1997, 323, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xie, J.; Jiang, Z.; Wang, B.; Wang, Y.; Hu, X. Shikonin and its analogs inhibit cancer cell glycolysis by targeting tumor pyruvate kinase-M2. Oncogene 2011, 30, 4297–4306. [Google Scholar] [CrossRef] [PubMed]
- Ibsen, K.H. Interrelationships and functions of the pyruvate kinase isozymes and their variant forms: A review. Cancer Res. 1977, 37, 341–353. [Google Scholar] [PubMed]
- Christofk, H.R.; Vander Heiden, M.G.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.D.; Schreiber, S.L.; Cantley, L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Spoden, G.A.; Mazurek, S.; Morandell, D.; Bacher, N.; Ausserlechner, M.J.; Jansen-Durr, P.; Eigenbrodt, E.; Zwerschke, W. Isotype-specific inhibitors of the glycolytic key regulator pyruvate kinase subtype M2 moderately decelerate tumor cell proliferation. Int. J. Cancer 2008, 123, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Spoden, G.A.; Rostek, U.; Lechner, S.; Mitterberger, M.; Mazurek, S.; Zwerschke, W. Pyruvate kinase isoenzyme M2 is a glycolytic sensor differentially regulating cell proliferation, cell size and apoptotic cell death dependent on glucose supply. Exp. Cell Res. 2009, 315, 2765–2774. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Brownlee, M. Hyperglycemia-induced reactive oxygen species increase expression of the receptor for advanced glycation end products (rage) and rage ligands. Diabetes 2010, 59, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liang, X.; Zheng, X.; Huang, H.; Chen, X.; Wu, K.; Wang, B.; Ma, S. Glo1 genetic amplification as a potential therapeutic target in hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 2079–2090. [Google Scholar] [PubMed]
- Shafie, A.; Xue, M.; Thornalley, P.J.; Rabbani, N. Copy number variation of glyoxalase I. Biochem. Soc. Trans. 2014, 42, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Birkenmeier, G.; Hemdan, N.Y.; Kurz, S.; Bigl, M.; Pieroh, P.; Debebe, T.; Buchold, M.; Thieme, R.; Wichmann, G.; Dehghani, F. Ethyl pyruvate combats human leukemia cells but spares normal blood cells. PLoS ONE 2016, in press. [Google Scholar] [CrossRef] [PubMed]
- Hollenbach, M.; Hintersdorf, A.; Huse, K.; Sack, U.; Bigl, M.; Groth, M.; Santel, T.; Buchold, M.; Lindner, I.; Otto, A.; et al. Ethyl pyruvate and ethyl lactate down-regulate the production of pro-inflammatory cytokines and modulate expression of immune receptors. Biochem. Pharmacol. 2008, 76, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Dong, A.; Fernandez-Ruiz, V.; Shan, J.; Kawa, M.; Martinez-Anso, E.; Prieto, J.; Qian, C. Blockade of WNT signaling inhibits angiogenesis and tumor growth in hepatocellular carcinoma. Cancer Res. 2009, 69, 6951–6959. [Google Scholar] [CrossRef] [PubMed]
- Hinck, L.; Nelson, W.J.; Papkoff, J. Wnt-1 modulates cell-cell adhesion in mammalian cells by stabilizing beta-catenin binding to the cell adhesion protein cadherin. J. Cell Biol. 1994, 124, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Lindner, I.; Hemdan, N.Y.; Buchold, M.; Huse, K.; Bigl, M.; Oerlecke, I.; Ricken, A.; Gaunitz, F.; Sack, U.; Naumann, A.; et al. α2-Macroglobulin inhibits the malignant properties of astrocytoma cells by impeding beta-catenin signaling. Cancer Res. 2010, 70, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Kimura, S.; Kuroda, J.; Ashihara, E.; Kawatani, M.; Osada, H.; Umezawa, K.; Yasui, E.; Imoto, M.; Tsuruo, T.; et al. Glyoxalase-I is a novel target against Bcr-Abl+ leukemic cells acquiring stem-like characteristics in a hypoxic environment. Cell Death Differ. 2010, 17, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Brouwers, O.; Niessen, P.M.; Ferreira, I.; Miyata, T.; Scheffer, P.G.; Teerlink, T.; Schrauwen, P.; Brownlee, M.; Stehouwer, C.D.; Schalkwijk, C.G. Overexpression of glyoxalase-I reduces hyperglycemia-induced levels of advanced glycation end products and oxidative stress in diabetic rats. J. Biol. Chem. 2011, 286, 1374–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, Y.; Inagi, R.; Miyata, T.; Nagai, R.; Arai, M.; Miyashita, M.; Itokawa, M.; Fujita, T.; Nangaku, M. Glyoxalase I retards renal senescence. Am. J. Pathol. 2011, 179, 2810–2821. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Rabbani, N.; Momiji, H.; Imbasi, P.; Anwar, M.M.; Kitteringham, N.; Park, B.K.; Souma, T.; Moriguchi, T.; Yamamoto, M.; et al. Transcriptional control of glyoxalase 1 by Nrf2 provides a stress-responsive defence against dicarbonyl glycation. Biochem. J. 2012, 443, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Birkenmeier, G.; Stegemann, C.; Hoffmann, R.; Gunther, R.; Huse, K.; Birkemeyer, C. Posttranslational modification of human glyoxalase 1 indicates redox-dependent regulation. PLoS ONE 2010, 5, e10399. [Google Scholar] [CrossRef] [PubMed]
- Nass, N.; Sel, S.; Ignatov, A.; Roessner, A.; Kalinski, T. Oxidative stress and glyoxalase I activity mediate dicarbonyl toxicity in MCF-7 mamma carcinoma cells and a tamoxifen resistant derivative. Biochim. Biophys. Acta 2016, 1860, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Inagi, R.; Kumagai, T.; Fujita, T.; Nangaku, M. The role of glyoxalase system in renal hypoxia. Adv. Exp. Med. Biol. 2010, 662, 49–55. [Google Scholar] [PubMed]
- Shi, D.Y.; Xie, F.Z.; Zhai, C.; Stern, J.S.; Liu, Y.; Liu, S.L. The role of cellular oxidative stress in regulating glycolysis energy metabolism in hepatoma cells. Mol. Cancer 2009, 8. [Google Scholar] [CrossRef] [PubMed]
- Rademakers, S.E.; Lok, J.; van der Kogel, A.J.; Bussink, J.; Kaanders, J.H. Metabolic markers in relation to hypoxia; staining patterns and colocalization of pimonidazole, HIF-1α, CAIX, LDH-5, GLUT-1, MCT1 and MCT4. BMC Cancer 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Weljie, A.M.; Jirik, F.R. Hypoxia-induced metabolic shifts in cancer cells: Moving beyond the warburg effect. Int. J. Biochem. Cell Biol. 2011, 43, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Van de Wetering, M.; Oving, I.; Muncan, V.; Pon Fong, M.T.; Brantjes, H.; van Leenen, D.; Holstege, F.C.; Brummelkamp, T.R.; Agami, R.; Clevers, H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003, 4, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Bigl, M.; Bleyl, A.D.; Zedlick, D.; Arendt, T.; Bigl, V.; Eschrich, K. Changes of activity and isozyme pattern of phosphofructokinase in the brains of patients with Alzheimer’s disease. J. Neurochem. 1996, 67, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Bergmeyer, H.U.; Bergmeyer, J.; Grassl, M. Methods of Enzymatic Analysis, 3rd ed.; Verlag Chemie: Weinheim, Germany, 1983; pp. 1–4. [Google Scholar]
- Mannervik, B.; Aronsson, A.C.; Tibbelin, G. Glyoxalase I from human erythrocytes. Methods Enzymol. 1982, 90, 535–541. [Google Scholar] [PubMed]
- Vander Jagt, D.L.; Robinson, B.; Taylor, K.K.; Hunsaker, L.A. Reduction of trioses by NADPH-dependent aldo-keto reductases. Aldose reductase, methylglyoxal, and diabetic complications. J. Biol. Chem. 1992, 267, 4364–4369. [Google Scholar] [PubMed]


| Activity in nkat/mg | MCF-7 | HEK 293 |
|---|---|---|
| PFK | 5.83 ± 0.33 | 1.76 ± 0.60 |
| HK | 0.83 ± 0.16 | 0.98 ± 0.16 |
| PK | 95.5 ± 10.0 | 28.3 ± 8.3 * |
| G6PDH | 14.8 ± 1.50 | 1.75 ± 0.3 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hutschenreuther, A.; Bigl, M.; Hemdan, N.Y.A.; Debebe, T.; Gaunitz, F.; Birkenmeier, G. Modulation of GLO1 Expression Affects Malignant Properties of Cells. Int. J. Mol. Sci. 2016, 17, 2133. https://doi.org/10.3390/ijms17122133
Hutschenreuther A, Bigl M, Hemdan NYA, Debebe T, Gaunitz F, Birkenmeier G. Modulation of GLO1 Expression Affects Malignant Properties of Cells. International Journal of Molecular Sciences. 2016; 17(12):2133. https://doi.org/10.3390/ijms17122133
Chicago/Turabian StyleHutschenreuther, Antje, Marina Bigl, Nasr Y. A. Hemdan, Tewodros Debebe, Frank Gaunitz, and Gerd Birkenmeier. 2016. "Modulation of GLO1 Expression Affects Malignant Properties of Cells" International Journal of Molecular Sciences 17, no. 12: 2133. https://doi.org/10.3390/ijms17122133






