Tamoxifen Treatment of Breast Cancer Cells: Impact on Hedgehog/GLI1 Signaling
Abstract
:1. Introduction
2. Results
2.1. Proliferation Assays
2.2. qRT–PCR Assays, GLI1 Variants, SMO and SHH Expression
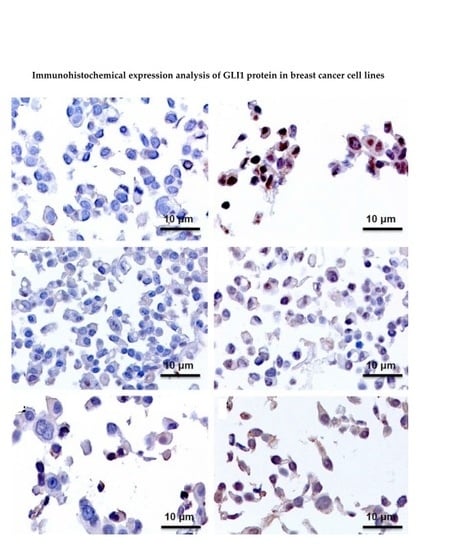
2.3. Immunohistochemistry Assays (IHC), Protein Expression of GLI1 in Nucleus and Cytoplasm
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Treatment of the Cell Lines with TAM
4.3. Proliferation Assays
4.4. RNA Isolation
4.5. Real-Time PCR
4.6. Immunohistochemistry Assays (IHC)
4.7. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Ramaswamy, B.; Lu, Y.; Teng, K.Y.; Nuovo, G.; Li, X.; Shapiro, C.L.; Majumder, S. Hedgehog signaling is a novel therapeutic target in tamoxifen-resistant breast cancer aberrantly activated by PI3K/AKT pathway. Cancer Res. 2012, 72, 5048–5059. [Google Scholar] [CrossRef] [PubMed]
- Takezaki, T.; Hide, T.; Takanaga, H.; Nakamura, H.; Kuratsu, J.I.; Kondo, T. Essential role of the Hedgehog signaling pathway in human glioma-initiating cells. Cancer Sci. 2011, 102, 1306–1312. [Google Scholar] [CrossRef] [PubMed]
- Kinzler, K.W.; Vogelstein, B. The Gli gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell. Biol. 1990, 10, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Kogerman, P.; Grimm, T.; Kogerman, L.; Krause, D.; Undén, A.B.; Sandstedt, B.; Toftgård, R.; Zaphiropoulos, P.G. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of gli-1. Nat. Cell Biol. 1999, 1, 312–319. [Google Scholar] [PubMed]
- Kasper, M.; Regl, G.; Frischauf, A.-M.; Aberger, F. Gli transcription factors: Mediators of oncogenic hedgehog signalling. Eur. J. Cancer 2006, 42, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, R.L.; Lo, H.-W. Hedgehog pathway and GLI1 isoforms in human cancer. Discov. Med. 2012, 13, 105–113. [Google Scholar] [PubMed]
- Shimokawa, T.; Tostar, U.; Lauth, M.; Palaniswamy, R.; Kasper, M.; Toftgård, R.; Zaphiropoulos, P.G. Novel human glioma-associated oncogene 1 (GLI1) splice variants reveal distinct mechanisms in the terminal transduction of the hedgehog signal. J. Biol. Chem. 2008, 283, 14345–14354. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.-W.; Zhu, H.; Cao, X.; Aldrich, A.; Ali-Osman, F. A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion. Cancer Res. 2009, 69, 6790–6798. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Gao, H.F.; Wang, Y.; Liu, F.; Tian, X.F.; Zhang, Y. Overexpression of Gli1 in cancer interstitial tissues predicts early relapse after radical operation of breast cancer. Chin. J. Cancer Res. 2012, 24, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Jordan, V.C. Tamoxifen: Catalyst for the change to targeted therapy. Eur. J. Cancer 2008, 44, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Osborne, C. Tamoxifen in the treatment of breast cancer. N. Engl. J. Med. 1998, 339, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Cronin-Fenton, D.P.; Damkier, P.; Lash, T.L. Metabolism and transport of tamoxifen in relation to its effectiveness: New perspectives on an ongoing controversy. Future Oncol. 2014, 10, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Takebe, N.; Lorusso, P. Targeting the Hedgehog pathway in cancer. Ther. Adv. Med. Oncol. 2010, 2, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Yauch, R.L.; Gould, S.E.; Scales, S.J.; Tang, T.; Tian, H.; Ahn, C.P.; Marshall, D.; Fu, L.; Januario, T.; Kallop, D.; et al. A paracrine requirement for hedgehog signalling in cancer. Nature 2008, 455, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Lauth, M.; Toftgård, R. Non-canonical activation of GLI transcription factors: Implications for targeted anti-cancer therapy. Cell Cycle 2007, 6, 2458–2463. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, D. Hedgehog signalling: Emerging evidence for non-canonical pathways. Cell Signal. 2009, 21, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Kameda, C.; Tanaka, H.; Yamasaki, A.; Nakamura, M.; Koga, K.; Sato, N.; Kubo, M.; Kuroki, S.; Tanaka, M.; Katano, M. The Hedgehog pathway is a possible therapeutic target for patients with estrogen receptor-negative breast cancer. Anticancer Res. 2009, 29, 871–879. [Google Scholar] [PubMed]
- Ten Haaf, A.; Bektas, N.; von Serenyi, S.; Losen, I.; Arweiler, E.C.; Hartmann, A.; Knüchel, R.; Dahl, E. Expression of the glioma-associated oncogene homolog (GLI) 1 in human breast cancer is associated with unfavourable overall survival. BMC Cancer 2009, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souzaki, M.; Kubo, M.; Kai, M.; Kameda, C.; Tanaka, H.; Taguchi, T.; Tanaka, M.; Onishi, H.; Katano, M. Hedgehog signaling pathway mediates the progression of non-invasive breast cancer to invasive breast cancer. Cancer Sci. 2011, 102, 373–381. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, S.A.; Machalek, D.A.; Shearer, R.F.; Millar, E.K.A.; Nair, R.; Schofield, P.; McLeod, D.; Cooper, C.L.; McNeil, C.M.; McFarland, A.; et al. Hedgehog overexpression is associated with stromal interactions and predicts for poor outcome in breast cancer. Cancer Res. 2011, 71, 4002–4014. [Google Scholar] [CrossRef] [PubMed]
- Narvaez, C.J.; Matthews, D.; LaPorta, E.; Simmons, K.M.; Beaudin, S.; Welsh, J. The impact of vitamin D in breast cancer: Genomics, pathways, metabolism. Front. Physiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Bijlsma, M.F.; Spek, C.A.; Zivkovic, D.; van de Water, S.; Rezaee, F.; Peppelenbosch, M.P. Repression of smoothened by patched-dependent (pro-) vitamin D3 secretion. PLoS Biol. 2006, 4, e232. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Okumura, T.; Tsunoda, S.; Sakai, Y.; Shimada, Y. Gli-1 expression is associated with lymph node metastasis and tumor progression in esophageal squamous cell carcinoma. Oncology 2006, 70, 378–389. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, R.; Nakano, Y.; Tao, L.; Koishi, K.; Matsumoto, T.; Sasako, M.; Tsujimura, T.; Hashimoto-Tamaoki, T.; Fujiwara, Y. Hedgehog signal activation in oesophageal cancer patients undergoing neoadjuvant chemo radiotherapy. Br. J. Cancer 2008, 98, 1670–1674. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-Q.; Russo, J. ERα-negative and triple negative breast cancer: Molecular features and potential therapeutic approaches. Biochim. Biophys. Acta 2009, 1796, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, L.; Zhang, G.; Zhang, L.; Chen, C. G protein-coupled receptor 30 in tumor development. Endocrine 2010, 38, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Girgert, R.; Emons, G.; Gründker, C. Inactivation of GPR30 reduces growth of triple-negative breast cancer cells: Possible application in targeted therapy. Breast Cancer Res. Treat. 2012, 134, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Prossnitz, E.R.; Arterburn, J.B.; Sklar, L.A. A G protein-coupled receptor for estrogen. Mol. Cell. Endocrinol. 2007, 265–266, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.; Liu, M.; Yang, F.; Luo, H.; Li, Z.; Tu, G.; Yang, G. GPR30 as an initiator of tamoxifen resistance in hormone-dependent breast cancer. Breast Cancer Res. 2013, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Levenson, A.S.; Jordan, V.C. MCF-7: The first hormone-responsive breast cancer cell line. Cancer Res. 1997, 57, 3071–3078. [Google Scholar] [PubMed]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13. [Google Scholar] [CrossRef] [PubMed]
- Infante, P.; Alfonsi, R.; Botta, B.; Mori, M.; di Marcotullio, L. Targeting GLI factors to inhibit the Hedgehog pathway. Trends Pharmacol. Sci. 2015, 36, 547–558. [Google Scholar] [CrossRef] [PubMed]







| Cell Line | Control | TAM 24 h | TAM 48 h | TAM 96 h |
|---|---|---|---|---|
| MCF7 | Negative | Positive (Nucleus and Cytoplasm) | Positive (Nucleus and Cytoplasm) | Positive (Nucleus and Cytoplasm) |
| T47D | Positive | Positive + (Nucleus and Cytoplasm) | Negative | Negative |
| ZR-75-1 | Negative | Positive (Nucleus) | Positive (Nucleus) | Positive (Nucleus) |
| BT474 | Negative | Positive (Nucleus) | Positive (Nucleus) | Positive (Nucleus) |
| SKBR3 | Positive | Positive + (Nucleus and Cytoplasm) | Positive + (Nucleus and Cytoplasm) | Positive + (Nucleus and Cytoplasm) |
| JIMT-1 | Negative | Negative | Negative | Positive + (Nucleus and Cytoplasm) |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villegas, V.E.; Rondón-Lagos, M.; Annaratone, L.; Castellano, I.; Grismaldo, A.; Sapino, A.; Zaphiropoulos, P.G. Tamoxifen Treatment of Breast Cancer Cells: Impact on Hedgehog/GLI1 Signaling. Int. J. Mol. Sci. 2016, 17, 308. https://doi.org/10.3390/ijms17030308
Villegas VE, Rondón-Lagos M, Annaratone L, Castellano I, Grismaldo A, Sapino A, Zaphiropoulos PG. Tamoxifen Treatment of Breast Cancer Cells: Impact on Hedgehog/GLI1 Signaling. International Journal of Molecular Sciences. 2016; 17(3):308. https://doi.org/10.3390/ijms17030308
Chicago/Turabian StyleVillegas, Victoria E., Milena Rondón-Lagos, Laura Annaratone, Isabella Castellano, Adriana Grismaldo, Anna Sapino, and Peter G. Zaphiropoulos. 2016. "Tamoxifen Treatment of Breast Cancer Cells: Impact on Hedgehog/GLI1 Signaling" International Journal of Molecular Sciences 17, no. 3: 308. https://doi.org/10.3390/ijms17030308