Chromosome 9p21 and ABCA1 Genetic Variants and Their Interactions on Coronary Heart Disease and Ischemic Stroke in a Chinese Han Population
Abstract
:1. Introduction
2. Results
2.1. Clinical Characteristics
2.2. Polymerase Chain Reaction (PCR) Products and Genotypes
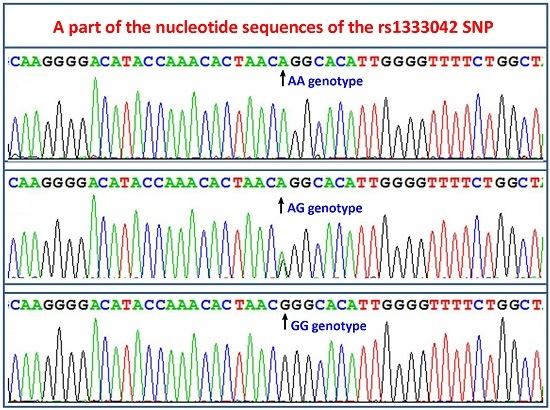
2.3. Nucleotide Sequences
2.4. Genotypic and Allelic Frequencies
2.5. Genotypes and the Risk of Coronary Heart Disease (CHD) and Ischemic Stroke (IS)
2.6. Linkage Disequilibrium (LD) Analyses
2.7. Gene-Environment Interactions on the Risk of CHD and IS
2.8. Gene–Gene Interactions on the Risk of CHD and IS
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Biochemical Measurements
4.3. Single Nucleotide Polymorphisms (SNP) Selection and Genotyping
4.4. Deoxyribonucleic Acid (DNA) Sequencing
4.5. Statistical Analyses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ABCA1 | Adenosine triphosphate-binding cassette transporter A1 |
| Apo | Apolipoprotein |
| BMI | Body mass index |
| CHD | Coronary heart disease |
| DNA | Deoxyribonucleic acid |
| GWASes | Genome-wide association studies |
| HDL-C | High-density lipoprotein cholesterol |
| IS | Ischemic stroke |
| LD | Linkage disequilibrium |
| LDL-C | Low-density lipoprotein cholesterol |
| MAF | Minor allele frequency |
| MDR | Multifactor dimensionality reduction |
| MRI | Magnetic resonance imaging |
| PCR | Polymerase chain reaction |
| RFLP | Restriction fragment length polymorphism |
| SNPs | Single nucleotide polymorphisms |
| TC | Total cholesterol |
| TG | Triglyceride |
| TOAST | Trial of Org 10172 in Acute Stroke Treatment |
References
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; de Ferranti, S.; Després, J.P.; Fullerton, H.J.; Howard, V.J.; et al. Heart disease and stroke statistics—2015 update: A report from the American Heart Association. Circulation 2015, 131, E29–E322. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Matsui, K.; Takeuchi, I.; Fujimaki, T. Association of genetic variants with coronary artery disease and ischemic stroke in a longitudinal population-based genetic epidemiological study. Biomed. Rep. 2015, 3, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Xu, Y.; Wang, X.; Wang, Q.; Zhang, L.; Tu, Y.; Yan, J.; Wang, W.; Hui, R.; Wang, C.Y.; et al. 9p21 is a shared susceptibility locus strongly for coronary artery disease and weakly for ischemic stroke in Chinese Han population. Circ. Cardiovasc. Genet. 2009, 2, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Lim, C.C.; Silver, L.E.; Welch, S.J.; Banning, A.P.; Rothwell, P.M. Familial history of stroke is associated with acute coronary syndromes in women. Circ. Cardiovasc. Genet. 2011, 4, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Sabater-Lleal, M.; Huang, J.; Chasman, D.; Naitza, S.; Dehghan, A.; Johnson, A.D.; Teumer, A.; Reiner, A.P.; Folkersen, L.; Basu, S.; et al. Multiethnic meta-analysis of genome-wide association studies in >100,000 subjects identifies 23 fibrinogen-associated Loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation 2013, 128, 1310–1324. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.G.; Melander, O.; Lövkvist, H.; Hedblad, B.; Engström, G.; Nilsson, P.; Carlson, J.; Berglund, G.; Norrving, B.; Lindgren, A. Common genetic variants on chromosome 9p21 confers risk of ischemic stroke: A large-scale genetic association study. Circ. Cardiovasc. Genet. 2009, 2, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Calling, S.; Ji, J.; Sundquist, J.; Sundquist, K.; Zöller, B. Shared and non-shared familial susceptibility of coronary heart disease, ischemic stroke, peripheral artery disease and aortic disease. Int. J. Cardiol. 2013, 168, 2844–2850. [Google Scholar] [CrossRef] [PubMed]
- Dichgans, M.; Malik, R.; König, I.R.; Rosand, J.; Clarke, R.; Gretarsdottir, S.; Thorleifsson, G.; Mitchell, B.D.; Assimes, T.L.; Levi, C.; et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: A genome-wide analysis of common variants. Stroke 2014, 45, 24–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, M.; Broeckel, U.; Holmer, S.; Baessler, A.; Hengstenberg, C.; Mayer, B.; Erdmann, J.; Klein, G.; Riegger, G.; Jacob, H.J.; et al. Distinct heritable patterns of angiographic coronary artery disease in families with myocardial infarction. Circulation 2005, 111, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Vargas, J.D.; Manichaikul, A.; Wang, X.Q.; Rich, S.S.; Rotter, J.I.; Post, W.S.; Polak, J.F.; Budoff, M.J.; Bluemke, D.A. Detailed analysis of association between common single nucleotide polymorphisms and subclinical atherosclerosis: The multi-ethnic study of atherosclerosis. Data Brief 2016, 7, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Marenberg, M.E.; Risch, N.; Berkman, L.F.; Floderus, B.; de Faire, U. Genetic susceptibility to death from coronary heart disease in a study of twins. N. Engl. J. Med. 1994, 330, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Peden, J.F.; Farrall, M. Thirty-five common variants for coronary artery disease: The fruits of much collaborative labour. Hum. Mol. Genet. 2011, 20, R198–R205. [Google Scholar] [CrossRef] [PubMed]
- Bevan, S.; Traylor, M.; Adib-Samii, P.; Malik, R.; Paul, N.L.; Jackson, C.; Farrall, M.; Rothwell, P.M.; Sudlow, C.; Dichgans, M.; et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke 2012, 43, 3161–3167. [Google Scholar] [CrossRef] [PubMed]
- Helgadottir, A.; Thorleifsson, G.; Manolescu, A.; Gretarsdottir, S.; Blondal, T.; Jonasdottir, A.; Jonasdottir, A.; Sigurdsson, A.; Baker, A.; Palsson, A.; et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 2007, 316, 1491–1493. [Google Scholar] [CrossRef] [PubMed]
- AbdulAzeez, S.; Al-Nafie, A.N.; Al-Shehri, A.; Borgio, J.F.; Baranova, E.V.; Al-Madan, M.S.; Al-Ali, R.A.; Al-Muhanna, F.; Al-Ali, A.; Al-Mansori, M.; et al. Intronic polymorphisms in the CDKN2B-AS1 gene are strongly associated with the risk of myocardial infarction and coronary artery disease in the Saudi population. Int. J. Mol. Sci. 2016, 17, 395. [Google Scholar] [CrossRef] [PubMed]
- Fedele, F.; Mancone, M.; Chilian, W.M.; Severino, P.; Canali, E.; Logan, S.; de Marchis, M.L.; Volterrani, M.; Palmirotta, R.; Guadagni, F. Role of genetic polymorphisms of ion channels in the pathophysiology of coronary microvascular dysfunction and ischemic heart disease. Basic Res. Cardiol. 2013, 108, 387. [Google Scholar] [CrossRef] [PubMed]
- Fedele, F.; Severino, P.; Bruno, N.; Stio, R.; Caira, C.; D’Ambrosi, A.; Brasolin, B.; Ohanyan, V.; Mancone, M. Role of ion channels in coronary microcirculation: A review of the literature. Future Cardiol. 2013, 9, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Scheffold, T.; Kullmann, S.; Huge, A.; Binner, P.; Ochs, H.R.; Schöls, W.; Thale, J.; Motz, W.; Hegge, F.J.; Stellbrink, C.; et al. Six sequence variants on chromosome 9p21.3 are associated with a positive family history of myocardial infarction: A multicenter registry. BMC Cardiovasc. Disord. 2011, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.D.; Biffi, A.; Rost, N.S.; Cortellini, L.; Furie, K.L.; Rosand, J. Chromosome 9p21 in ischemic stroke: Population structure and meta-analysis. Stroke 2010, 41, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Gschwendtner, A.; Bevan, S.; Cole, J.W.; Plourde, A.; Matarin, M.; Ross-Adams, H.; Meitinger, T.; Wichmann, E.; Mitchell, B.D.; Furie, K.; et al. Sequence variants on chromosome 9p21.3 confer risk for atherosclerotic stroke. Ann. Neurol. 2009, 65, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Hamon, Y.; Chimini, G. The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res. 2001, 42, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Remaley, A.T.; Rust, S.; Rosier, M.; Knapper, C.; Naudin, L.; Broccardo, C.; Peterson, K.M.; Koch, C.; Arnould, I.; Prades, C.; et al. Human ATP-binding cassette transporter 1 (ABC1): Genomic organization and identification of the genetic defect in the original Tangier disease kindred. Proc. Natl. Acad. Sci. USA 1999, 96, 12685–12690. [Google Scholar] [CrossRef] [PubMed]
- Frikke-Schmidt, R.; Nordestgaard, B.G.; Stene, M.C.; Sethi, A.A.; Remaley, A.T.; Schnohr, P.; Grande, P.; Tybjaerg-Hansen, A. Association of loss-of-function mutations in the ABCA1 gene with high-density lipoprotein cholesterol levels and risk of ischemic heart disease. JAMA 2008, 299, 2524–2532. [Google Scholar] [CrossRef] [PubMed]
- Porchay-Baldérelli, I.; Péan, F.; Emery, N.; Maimaitiming, S.; Bellili, N.; Travert, F.; Mohammedi, K.; Roussel, R.; Marre, M.; Fumeron, F.; et al. Relationships between common polymorphisms of adenosine triphosphate-binding cassette transporter A1 and high-density lipoprotein cholesterol and coronary heart disease in a population with type 2 diabetes mellitus. Metabolism 2009, 58, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Xue, X.H.; Lin, Y.; Fang, L.; Murong, S.; Wu, Z.Y. The R219K polymorphism in the ATP-binding cassette transporter 1 gene has a protective effect on atherothrombotic cerebral infarction in Chinese Han ethnic population. Neurobiol. Aging. 2010, 31, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Kakko, S.; Kelloniemi, J.; von Rohr, P.; Hoeschele, I.; Tamminen, M.; Brousseau, M.E.; Kesäniemi, Y.A.; Savolainen, M.J. ATP-binding cassette transporter A1 locus is not a major determinant of HDL-C levels in a population at high risk for coronary heart disease. Atherosclerosis 2003, 166, 285–290. [Google Scholar] [CrossRef]
- Pasdar, A.; Yadegarfar, G.; Cumming, A.; Whalley, L.; St Clair, D.; MacLeod, M.J. The effect of ABCA1 gene polymorphisms on ischaemic stroke risk and relationship with lipid profile. BMC Med. Genet. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Yumnam, S.; Basu, T.; Ghosh, A.; Garg, G.; Karthikeyan, G.; Sengupta, S. Association of polymorphisms in 9p21 region with CAD in North Indian population: Replication of SNPs identified through GWAS. Clin. Genet. 2011, 79, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.F.; Tsai, P.C.; Liao, Y.C.; Lin, T.H.; Tai, C.T.; Juo, S.H.; Lin, R.T. Chromosome 9p21 genetic variants are associated with myocardial infarction but not with ischemic stroke in a Taiwanese population. J. Investig. Med. 2011, 59, 926–930. [Google Scholar] [PubMed]
- Golabgir Khademi, K.; Foroughmand, A.M.; Galehdari, H.; Yazdankhah, S.; Pourmahdi Borujeni, M.; Shahbazi, Z.; Dinarvand, P. Association study of rs1333040 and rs1004638 polymorphisms in the 9p21 locus with coronary artery disease in Southwest of Iran. Iran Biomed. J. 2016, 20, 122–127. [Google Scholar] [PubMed]
- Zanetti, D.; Carreras-Torres, R.; Esteban, E.; Via, M.; Moral, P. Potential signals of natural selection in the top risk loci for coronary artery disease: 9p21 and 10q11. PLoS ONE 2015, 10, e0134840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, L.; Ma, J.; Qi, Q.; Hartiala, J.; Allayee, H.; Campos, H. Genetic risk score and risk of myocardial infarction in Hispanics. Circulation 2011, 123, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Lee, B.S.; Shin, D.J.; Woo Park, K.; Shin, Y.A.; Joong Kim, K.; Heo, L.; Young Lee, J.; Kyoung Kim, Y.; Jin Kim, Y.; et al. A genome-wide association study of a coronary artery disease risk variant. J. Hum. Genet. 2013, 58, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Liu, X.; Zhang, Y.; Yu, L.; Zhang, F.; Liu, L.; Cai, J.; Yang, X.; Wang, X. Genetic variants associated with myocardial infarction and the risk factors in Chinese population. PLoS ONE 2014, 9, e86332. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, R.; Abe, S.; Tokoro, F.; Arai, M.; Noda, T.; Watanabe, S.; Horibe, H.; Fujimaki, T.; Oguri, M.; Kato, K.; et al. Association of six genetic variants with myocardial infarction. Int. J. Mol. Med. 2015, 35, 1451–1459. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.H.; Low, P.S.; Tan, Y.S.; Tong, M.C.; Saha, N.; Yang, H.; Heng, C.K. ABCA1 gene polymorphisms and their associations with coronary artery disease and plasma lipids in males from three ethnic populations in Singapore. Hum. Genet. 2003, 113, 106–117. [Google Scholar] [PubMed]
- Kyriakou, T.; Pontefract, D.E.; Viturro, E.; Hodgkinson, C.P.; Laxton, R.C.; Bogari, N.; Cooper, G.; Davies, M.; Giblett, J.; Day, I.N.; et al. Functional polymorphism in ABCA1 influences age of symptom onset in coronary artery disease patients. Hum. Mol. Genet. 2007, 16, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Sheĭdina, A.M.; Pchelina, S.N.; Demidova, D.V.; Rodygina, T.I.; Taraskina, A.E.; Toperverg, O.B.; Berkovich, O.A.; Demina, E.V.; Shvarts, E.I.; Pogoda, T.V.; et al. Allele frequency analysis of four single nucleotide polymorphisms locating in promoter and 5’-untranslated regions of ABCAI gene in young men—Survivors from myocardial infarction. Kardiologiia 2004, 44, 40–45. [Google Scholar] [PubMed]
- Zwarts, K.Y.; Clee, S.M.; Zwinderman, A.H.; Engert, J.C.; Singaraja, R.; Loubser, O.; James, E.; Roomp, K.; Hudson, T.J.; Jukema, J.W.; et al. ABCA1 regulatory variants influence coronary artery disease independent of effects on plasma lipid levels. Clin. Genet. 2002, 61, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Regieli, J.J.; Doevendans, P.A.; Grobbee, D.E.; Zwinderman, A.H.; van der Graaf, Y.; Kastelein, J.J.; Jukema, J.W. ABCA1 impacts athero-thrombotic risk and 10-year survival in a contemporary secondary prevention setting. Atherosclerosis 2011, 218, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Heckman, M.G.; Soto-Ortolaza, A.I.; Diehl, N.N.; Rayaprolu, S.; Brott, T.G.; Wszolek, Z.K.; Meschia, J.F.; Ross, O.A. Genetic variants associated with myocardial infarction in the PSMA6 gene and Chr9p21 are also associated with ischaemic stroke. Eur. J. Neurol. 2013, 20, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Lövkvist, H.; Sjögren, M.; Höglund, P.; Engström, G.; Jern, C.; Olsson, S.; Smith, J.G.; Hedblad, B.; Andsberg, G.; Delavaran, H.; et al. Are 25 SNPs from the CARDIoGRAM study associated with ischaemic stroke? Eur. J. Neurol. 2013, 20, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zhang, Y.; Maimaiti, Y.; Feng, Y.; Sun, J.; Zhuang, J.; Zeng, L.; Fu, Y. Variants on chromosome 9p21 confer risks of noncardioembolic cerebral infarction and carotid plaque in the Chinese Han population. J. Atheroscler. Thromb. 2015, 22, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.C.; Anderson, C.D.; Bione, S.; Keene, K.; Maguire, J.M.; Nalls, M.; Rasheed, A.; Zeginigg, M.; Attia, J.; Baker, R.; et al. Are myocardial infarction-associated single-nucleotide polymorphisms associated with ischemic stroke? Stroke 2012, 43, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Do, R.; Xie, C.; Zhang, X.; Männistö, S.; Harald, K.; Islam, S.; Bailey, S.D.; Rangarajan, S.; McQueen, M.J.; Diaz, R.; et al. The effect of chromosome 9p21 variants on cardiovascular disease may be modified by dietary intake: Evidence from a case/control and a prospective study. PLoS Med. 2011, 8, e1001106. [Google Scholar] [CrossRef] [PubMed]
- Hamrefors, V.; Hedblad, B.; Hindy, G.; Smith, J.G.; Almgren, P.; Engström, G.; Sjögren, M.; Gränsbo, K.; Orho-Melander, M.; Melander, O. Smoking modifies the associated increased risk of future cardiovascular disease by genetic variation on chromosome 9p21. PLoS ONE 2014, 9, e85893. [Google Scholar] [CrossRef] [PubMed]
- Leung Yinko, S.S.; Thanassoulis, G.; Stark, K.D.; Avgil Tsadok, M.; Engert, J.C.; Pilote, L.; GENESIS-PRAXY Investigators. ω-3 fatty acids and the genetic risk of early onset acute coronary syndrome. Nutr. Metab. Cardiovasc. Dis. 2014, 24, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.L.; Yin, R.X.; Wu, D.F.; Miao, L.; Aung, L.H.; Hu, X.J.; Li, Q.; Yan, T.T.; Lin, W.X.; Pan, S.L. Genetic variant of V825I in the ATP-binding cassette transporter A1 gene and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 2011, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Hinohara, K.; Nakajima, T.; Takahashi, M.; Hohda, S.; Sasaoka, T.; Nakahara, K.; Chida, K.; Sawabe, M.; Arimura, T.; Sato, A.; et al. Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J. Hum. Genet. 2008, 53, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Rivera, N.V.; Carreras-Torres, R.; Roncarati, R.; Viviani-Anselmi, C.; De Micco, F.; Mezzelani, A.; Koch, W.; Hoppmann, P.; Kastrati, A.; Stewart, A.F.; et al. Assessment of the 9p21.3 locus in severity of coronary artery disease in the presence and absence of type 2 diabetes. BMC Med. Genet. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Adams, H.P., Jr.; Bendixen, B.H.; Kappelle, L.J.; Biller, J.; Love, B.B.; Gordon, D.L.; Marsh, E.E., 3rd. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 1993, 24, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qiu, Q.; Tan, L.L.; Liu, T.; Deng, X.Q.; Chen, Y.M.; Chen, W.; Yu, X.Q.; Hu, B.J.; Chen, W.Q. Prevalence and determinants of diabetes and impaired fasting glucose among urban community-dwelling adults in Guangzhou, China. Diabetes Metab. 2009, 35, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.X.; Aung, L.H.; Long, X.J.; Yan, T.T.; Cao, X.L.; Huang, F.; Wu, J.Z.; Yang, D.Z.; Lin, W.X.; Pan, S.L. Interactions of several genetic polymorphisms and alcohol consumption on blood pressure levels. Biofactors 2015, 41, 339–351. [Google Scholar] [PubMed]
- Zhou, B.F.; Cooperative Meta-Analysis Group of the Working Group on Obesity in China. Predictive values of body mass index and waist circumference for risk factors of certain related diseases in Chinese adults—Study on optimal cut-off points of body mass index and waist circumference in Chinese adults. Biomed. Environ. Sci. 2002, 15, 83–96. [Google Scholar] [PubMed]
- Wildman, R.P.; Gu, D.; Reynolds, K.; Duan, X.; He, J. Appropriate body mass index and waist circumference cutoffs for categorization of overweight and central adiposity among Chinese adults. Am. J. Clin. Nutr. 2004, 80, 1129–1136. [Google Scholar] [PubMed]
- Aung, L.H.; Yin, R.X.; Wu, D.F.; Wang, W.; Liu, C.W.; Pan, S.L. Association of the variants in the BUD13-ZNF259 genes and the risk of hyperlipidaemia. J. Cell. Mol. Med. 2014, 18, 1417–1428. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Yin, R.X.; Huang, F.; Yao, L.M.; Lin, W.X.; Pan, S.L. Association between the DOCK7, PCSK9 and GALNT2 gene polymorphisms and serum lipid levels. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.D.; Hahn, L.W.; Roodi, N.; Bailey, L.R.; Dupont, W.D.; Parl, F.F.; Moore, J.H. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001, 69, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Coffey, C.S.; Hebert, P.R.; Ritchie, M.D.; Krumholz, H.M.; Gaziano, J.M.; Ridker, P.M.; Brown, N.J.; Vaughan, D.E.; Moore, J.H. An application of conditional logistic regression and multifactor dimensionality reduction for detecting gene–gene interactions on risk of myocardial infarction: The importance of model validation. BMC Bioinform. 2004, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, R.W.; Kaplan, N.L. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet. Epidemiol. 2002, 23, 221–233. [Google Scholar] [CrossRef] [PubMed]



| Parameter | Control | CHD | IS | p1 | p2 |
|---|---|---|---|---|---|
| Number | 541 | 565 | 569 | – | – |
| Male, n (%) | 346 (64.0) | 394 (69.7) | 396 (69.6) | 0.051 | 0.053 |
| Age, years | 61.39 ± 11.41 | 62.31 ± 10.53 | 62.53 ± 11.92 | 0.079 | 0.051 |
| Body mass index, kg/m2 | 22.49 ± 2.90 | 23.98 ± 3.21 | 23.50 ± 3.60 | 0.000 | 0.000 |
| Cigarette smoking, n (%) | 217 (40.1) | 244 (43.2) | 258 (45.7) | 0.300 | 0.078 |
| Diabetes, n (%) | 19 (3.5) | 129 (22.8) | 305 (53.98) | 0.000 | 0.000 |
| Hypertension, n (%) | 123 (22.7) | 282 (49.9) | 326 (57.3) | 0.000 | 0.000 |
| Hyperlipidemia, n (%) | 229 (42.3) | 277 (49.0) | 84 (14.9) | 0.025 | 0.000 |
| Total cholesterol, mmol/L | 4.84 ± 1.07 | 4.55 ± 1.24 | 4.52 ± 1.16 | 0.002 | 0.000 |
| Triglyceride, mmol/L | 1.01 (0.69) | 1.34 (0.93) | 1.36 (0.94) | 0.000 | 0.000 |
| HDL-C, mmol/L | 1.85 ± 0.51 | 1.14 ± 0.34 | 1.24 ± 0.41 | 0.000 | 0.000 |
| LDL-C, mmol/L | 2.77 ± 0.82 | 2.73 ± 1.04 | 2.67 ± 0.91 | 0.511 | 0.071 |
| Apolipoprotein (Apo)A1, g/L | 1.37 ± 0.28 | 1.05 ± 0.55 | 1.05 ± 0.55 | 0.000 | 0.000 |
| ApoB, g/L | 0.89 ± 0.21 | 0.90 ± 0.28 | 0.90 ± 0.26 | 0.344 | 0.431 |
| ApoA1/ApoB | 1.62 ± 0.55 | 1.40 ± 2.51 | 1.19 ± 0.60 | 0.042 | 0.000 |
| SNP/Group | Genotype | Allele | pHWE | ||||
|---|---|---|---|---|---|---|---|
| N | AA | AB | BB | A | B | ||
| rs1333040 (C>T) | |||||||
| Control | 541 | 60 (11.1) | 225 (41.6) | 256 (47.3) | 345 (31.9) | 737 (68.1) | 0.322 |
| CHD | 565 | 39 (6.9) | 232 (41.1) | 294 (52.0) | 310 (27.4) | 820 (72.6) | 0.457 |
| IS | 569 | 55 (9.7) | 247 (43.4) | 267 (46.9) | 357 (31.4) | 781 (68.6) | 0.846 |
| pCHD | 0.035 | 0.022 | |||||
| pIS | 0.681 | 0.794 | |||||
| rs1333042 (A>G) | |||||||
| Control | 541 | 67 (12.4) | 219 (40.5) | 227 (42.0) | 353 (32.6) | 729 (67.4) | 0.941 |
| CHD | 565 | 43 (7.60) | 227 (40.2) | 255 (59.2) | 313 (27.7) | 817 (72.3) | 0.065 |
| IS | 569 | 56 (9.7) | 243 (42.7) | 270 (47.6) | 355 (31.2) | 783 (68.8) | 0.066 |
| pCHD | 0.021 | 0.011 | |||||
| pIS | 0.376 | 0.470 | |||||
| rs4977574 (A>G) | |||||||
| Control | 541 | 152 (28.1) | 255 (47.1) | 134 (24.8) | 559 (51.7) | 523 (48.3) | 0.191 |
| CHD | 565 | 117 (20.7) | 272 (48.1) | 176 (31.2) | 506 (44.8) | 624 (55.2) | 0.527 |
| IS | 569 | 121 (21.3) | 276 (48.5) | 172 (30.2) | 518 (45.5) | 620 (54.5) | 0.599 |
| pCHD | 0.006 | 0.001 | |||||
| pIS | 0.015 | 0.004 | |||||
| rs2066715 (A>G) | |||||||
| Control | 541 | 97 (17.9) | 266 (49.2) | 178 (32.9) | 460 (42.5) | 622 (57.5) | 0.891 |
| CHD | 565 | 101 (17.9) | 298 (52.7) | 166 (29.4) | 500 (44.2) | 630 (55.8) | 0.101 |
| IS | 569 | 116 (20.4) | 279 (49.0) | 174 (30.6) | 627 (55.1) | 511 (44.9) | 0.823 |
| pCHD | 0.408 | 0.411 | |||||
| pIS | 0.510 | 0.256 | |||||
| rs2740483 (C>G) | |||||||
| Control | 541 | 31 (5.70) | 183 (33.8) | 327 (60.4) | 245 (22.6) | 837 (77.4) | 0.423 |
| CHD | 565 | 17 (2.8) | 175 (31.0) | 373 (66.2) | 209 (18.5) | 921 (81.5) | 0.516 |
| IS | 569 | 9 (1.6) | 152 (26.7) | 408 (71.7) | 170 (14.9) | 969 (85.1) | 0.223 |
| pCHD | 0.034 | 0.016 | |||||
| pIS | 0.000 | 0.000 | |||||
| SNP/Model | Ref. Genotype | Effect Genotype | CHD (OR 95% CI) | p | IS (OR 95% CI) | p |
|---|---|---|---|---|---|---|
| rs1333040 | ||||||
| Codominant | CC | CT | 1.74 (1.06–2.86) | 0.030 | 1.06 (0.69–1.65) | 0.781 |
| TT | 1.84 (1.13–3.01) | 0.014 | 1.13 (0.72–1.75) | 0.598 | ||
| Dominant | CC | CT/TT | 1.79 (1.12–2.88) | 0.016 | 1.09 (0.72–1.66) | 0.677 |
| Recessive | CC/CT | TT | 1.17 (0.90–1.52) | 0.244 | 0.97 (0.75–1.25) | 0.798 |
| Overdominant | CC/TT | CT | 1.03 (0.79–1.35) | 0.820 | 1.07 (0.83–1.39) | 0.606 |
| Log-additive | 1.23 (1.00–1.50) | 0.049 | 1.00 (0.83–1.21) | 0.994 | ||
| rs1333042 | ||||||
| Codominant | AA | AG | 1.73 (1.06–2.80) | 0.027 | 1.17 (0.76–1.79) | 0.483 |
| GG | 1.92 (1.20–3.08) | 0.007 | 1.21 (0.78–1.87) | 0.395 | ||
| Dominant | AA | AG/GG | 1.83 (1.16–2.89) | 0.010 | 1.19 (0.79–1.79) | 0.418 |
| Recessive | AA/AG | GG | 1.24 (0.95–1.61) | 0.115 | 1.00 (0.78–1.30) | 0.974 |
| Overdominant | AA/GG | AG | 1.00 (0.76–1.30) | 0.982 | 1.07 (0.82–1.38) | 0.629 |
| Log-additive | 1.27 (1.04–1.56) | 0.018 | 1.04 (0.86–1.26) | 0.689 | ||
| rs4977574 | ||||||
| Condominat | AA | AG | 1.44 (1.04–2.02) | 0.029 | 1.44 (1.05–1.98) | 0.025 |
| GG | 1.64 (1.14–2.37) | 0.008 | 1.63 (1.14–2.33) | 0.007 | ||
| Dominant | AA | AG/GG | 1.52 (1.11–2.07) | 0.009 | 1.51 (1.12–2.03) | 0.007 |
| Recessive | AA/AG | GG | 1.28 (0.96–1.72) | 0.095 | 1.28 (0.96–1.70) | 0.092 |
| Overdominant | AA/GG | AG | 1.11 (0.85–1.45) | 0.446 | 1.11 (0.86–1.43) | 0.427 |
| Log-additive | 1.27 (1.06–1.53) | 0.010 | 1.27 (1.06–1.52) | 0.008 | ||
| rs2066715 | ||||||
| Codominant | GG | AG | 1.00 (0.67–1.48) | 0.994 | 1.10 (0.76–1.59) | 0.623 |
| AA | 1.19 (0.88–1.61) | 0.260 | 1.00 (0.75–1.34) | 0.992 | ||
| Dominant | GG | AG/AA | 1.14 (0.85–1.51) | 0.382 | 1.03 (0.78–1.35) | 0.849 |
| Recessive | GG/AG | AA | 0.90 (0.63–1.27) | 0.537 | 1.00 (0.79–1.52) | 0.580 |
| Overdominant | GG/AA | AG | 1.19 (0.91–1.55) | 0.199 | 0.97 (0.75–1.25) | 0.801 |
| Log-additive | 1.02 (0.85–1.24) | 0.806 | 1.04 (0.87–1.25) | 0.663 | ||
| rs2740483 | ||||||
| Codominant | GG | GC | 0.82 (0.61–1.08) | 0.159 | 0.64 (0.48–0.85) | 0.002 |
| CC | 0.43 (0.22–0.83) | 0.012 | 0.22 (0.10–0.49) | 0.000 | ||
| Dominant | GG | CG/CC | 0.76 (0.58–0.99) | 0.044 | 0.58 (0.44–0.76) | 0.000 |
| Recessive | GG/CG | CC | 0.46 (0.24–0.89) | 0.020 | 0.25 (0.11–0.56) | 0.001 |
| Overdominant | GG/CC | CG | 0.86 (0.65–1.14) | 0.305 | 0.69 (0.52–0.91) | 0.009 |
| Log-additive | 0.74 (0.59–0.94) | 0.011 | 0.58 (0.46–0.73) | 0.000 | ||
| Factor | rs1333042 (pCHD) | rs4977574 (pCHD/pIS) | rs2740483 (pCHD/pIS) |
|---|---|---|---|
| Sex (male vs. female) | 0.967 | 0.729/0.940 | 0.560/0.128 |
| Age (≤60 vs. >60 year) | 0.444 | 0.022/0.216 | 0.213/0.490 |
| BMI (≤24 vs. >24 kg/m2) | 0.565 | 0.720/0.824 | 0.198/0.035 |
| Smoking (yes vs. no) | 0.099 | 0.882/0.299 | 0.839/0.539 |
| Hypertension (yes vs. no) | 0.815 | 0.876/0.990 | 0.837/0.331 |
| Diabetes (yes vs. no) | NA | 0.119/NA | 0.192/0.159 |
| Hyperlipidemia (yes vs. no) | 0.945 | 0.125/0.430 | 0.797/0.607 |
| Model | MDR Analysis | LR Analysis | |||||
|---|---|---|---|---|---|---|---|
| Traing Bal.Acc | Testing Bal.Acc | CVC | p | Wald | OR (95% CI) | p | |
| rs4977574 | 0.538 | 0.518 | 9/10 | 0.377 | – | – | – |
| A-B | 0.574 | 0.566 | 10/10 | 0.011 | 15.98 | 1.69 (1.31–2.19) | 0.000 |
| A-B-C | 0.601 | 0.536 | 7/10 | 0.054 | 7.52 | 1.16 (1.04–1.73) | 0.006 |
| A-B-C-D | 0.637 | 0.601 | 10/10 | 0.001 | 5.97 | 1.08 (1.02–1.14) | 0.015 |
| Model | MDR Analysis | LR Analysis | |||||
|---|---|---|---|---|---|---|---|
| Traing Bal.Acc | Testing Bal.Acc | CVC | p | Wald | OR (95% CI) | p | |
| rs2740483 | 0.557 | 0.555 | 10/10 | 0.001 | - | - | - |
| A-B | 0.603 | 0.602 | 10/10 | 0.001 | 20.56 | 1.80 (1.40–2.32) | 0.000 |
| A-B-C | 0.624 | 0.594 | 8/10 | 0.011 | 13.56 | 1.19 (0.90–1.27) | 0.000 |
| A-B-C-rs2740483 | 0.654 | 0.602 | 10/10 | 0.001 | 0.22 | 1.02 (0.94–1.40) | 0.640 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, X.-L.; Yin, R.-X.; Huang, F.; Wu, J.-Z.; Chen, W.-X. Chromosome 9p21 and ABCA1 Genetic Variants and Their Interactions on Coronary Heart Disease and Ischemic Stroke in a Chinese Han Population. Int. J. Mol. Sci. 2016, 17, 586. https://doi.org/10.3390/ijms17040586
Cao X-L, Yin R-X, Huang F, Wu J-Z, Chen W-X. Chromosome 9p21 and ABCA1 Genetic Variants and Their Interactions on Coronary Heart Disease and Ischemic Stroke in a Chinese Han Population. International Journal of Molecular Sciences. 2016; 17(4):586. https://doi.org/10.3390/ijms17040586
Chicago/Turabian StyleCao, Xiao-Li, Rui-Xing Yin, Feng Huang, Jin-Zhen Wu, and Wu-Xian Chen. 2016. "Chromosome 9p21 and ABCA1 Genetic Variants and Their Interactions on Coronary Heart Disease and Ischemic Stroke in a Chinese Han Population" International Journal of Molecular Sciences 17, no. 4: 586. https://doi.org/10.3390/ijms17040586