The Distribution of Lectins across the Phylum Nematoda: A Genome-Wide Search
Abstract
:1. Introduction
2. Results
2.1. Selection of Lectin-Like Domains
2.2. Predicting Carbohydrate Binding Properties
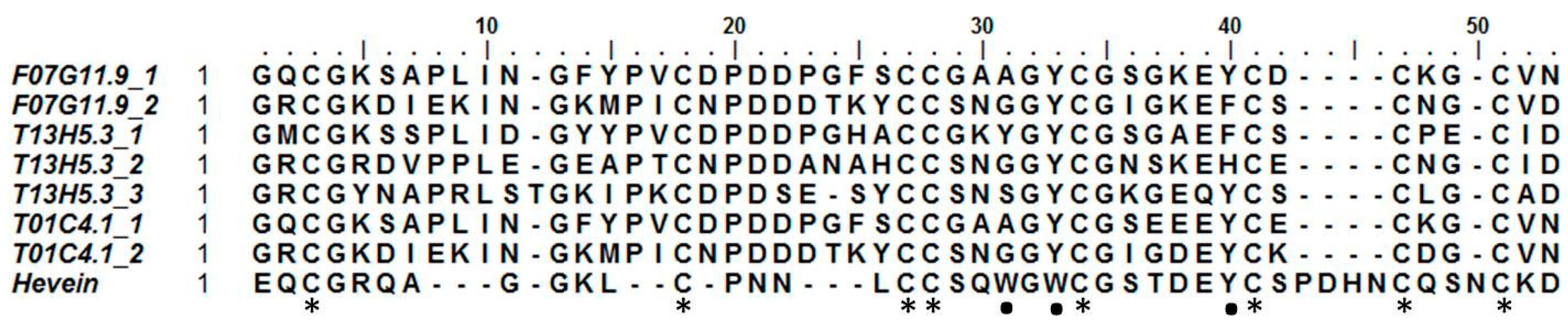
2.3. Hevein-Type Lectins
2.4. C-Type Lectin-Like Domains
2.5. General Overview
3. Discussion
4. Materials and Methods
4.1. Lectin-Like Domain Discovery
4.2. Structural Classification
4.3. Conserved Sequences
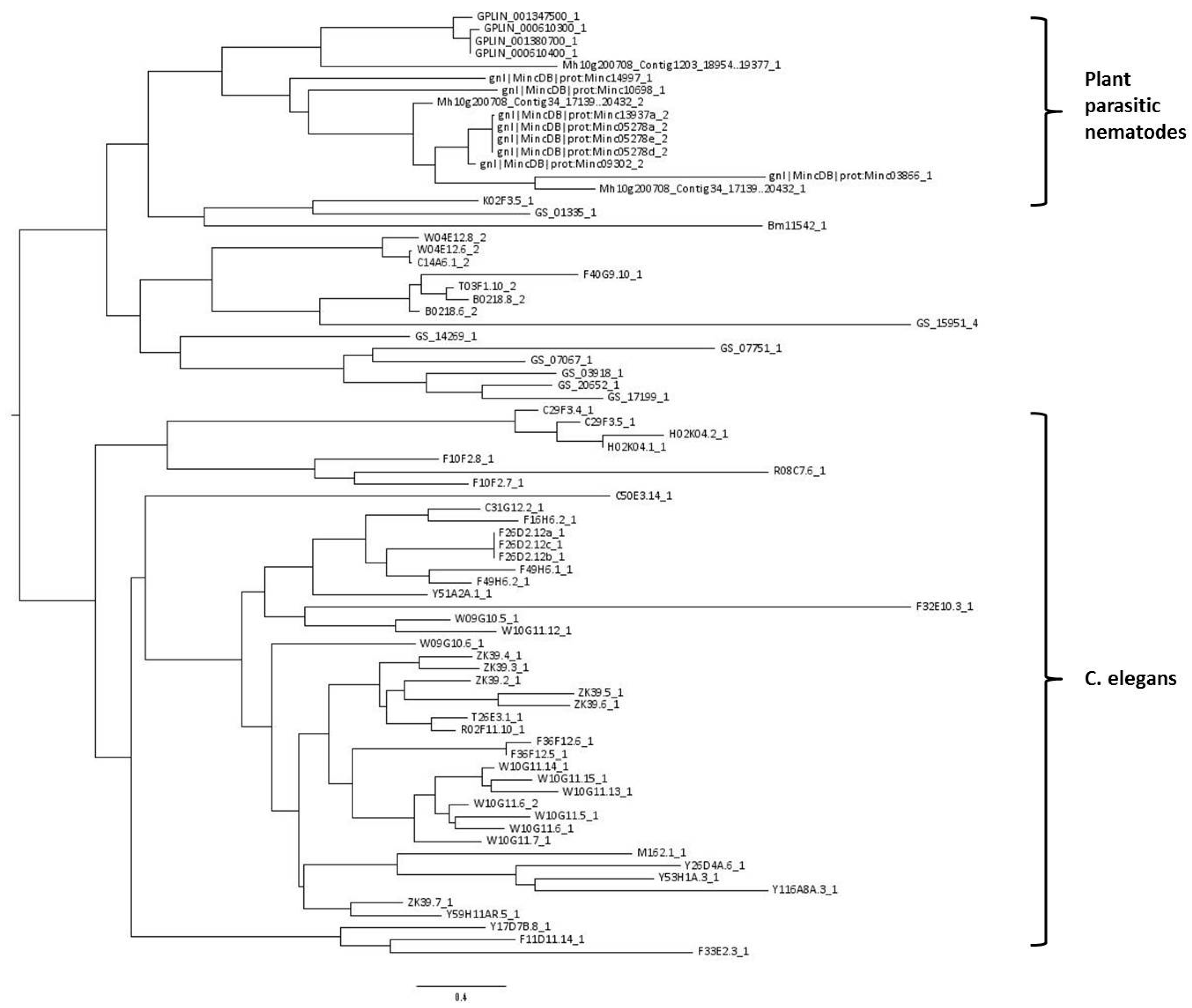
4.4. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CTLD | C-type lectin-like domain |
| CE | C. elegans |
| BM | B. malayi |
| AS | A. suum |
| MI | M. incognita |
| MH | M. hapla |
| MG | M. graminicola |
| BX | B. xylophilus |
| GP | G. pallida |
| HA | H. avenae |
| HG | H. glycines |
| HO | H. oryzae |
| PC | P. coffeae |
| PT | P. thornei |
| Trp | Tryptophan |
| Tyr | Tyrosine |
| GHF | Glycosyl hydrolase family |
| HMM | Hidden Markov model |
| EDEM | ER degradation-enhancing α-mannosidase-like protein |
References
- Kilpatrick, D.C. Animal lectins: A historical introduction and overview. Biochim. Biophys. Acta 2002, 1572, 187–197. [Google Scholar] [CrossRef]
- Peumans, W.J.; van Damme, E.J. Lectins as plant defense proteins. Plant Physiol. 1995, 109, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Sharon, N.; Lis, H. History of lectins: From hemagglutinins to biological recognition molecules. Glycobiology 2004, 14, 53R–62R. [Google Scholar] [CrossRef] [PubMed]
- Stillmark, H. About Ricin, a Toxic Enzyme from the Seeds of Ricinus communis L. and Some Other Euphorbiaceae; Medical School of Dorpat: Tartu, Estonia, 1888. [Google Scholar]
- Watkins, W.M.; Morgan, W.T. Neutralization of the anti-H agglutinin in eel serum by simple sugars. Nature 1952, 169, 825–826. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.; Lannoo, N.; Peumans, W.J. Plant lectins. Adv. Bot. Res. 2008, 48, 107–209. [Google Scholar]
- Van Damme, E.J. History of plant lectin research. Methods Mol. Biol. 2014, 1200, 3–13. [Google Scholar] [PubMed]
- Stockert, R.J.; Morell, A.G.; Scheinberg, I.H. Mammalian hepatic lectin. Science 1974, 186, 365–366. [Google Scholar] [CrossRef] [PubMed]
- Flexner, S.; Noguchi, H. Snake venom in relation to haemolysis, bacteriolysis and toxicity. J. Exp. Med. 1902, 6, 277–301. [Google Scholar] [CrossRef] [PubMed]
- Loris, R. Principles of structures of animal and plant lectins. Biochim. Biophys. Acta 2002, 1572, 198–208. [Google Scholar] [CrossRef]
- Drickamer, K. Two distinct classes of carbohydrate-recognition domains in animal lectins. J. Biol. Chem. 1988, 263, 9557–9560. [Google Scholar] [PubMed]
- Dodd, R.B.; Drickamer, K. Lectin-like proteins in model organisms: Implications for evolution of carbohydrate-binding activity. Glycobiology 2001, 11, 71R–79R. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.E.; Drickamer, K. Convergent and divergent mechanisms of sugar recognition across kingdoms. Curr. Opin. Struct. Biol. 2014, 28, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, J.; Ubukata, T.; Kasai, K. Purification and molecular characterization of a novel 16-kDa galectin from the nematode Caenorhabditis elegans. J. Biol. Chem. 1996, 271, 2497–2505. [Google Scholar] [CrossRef] [PubMed]
- Loukas, A.; Mullin, N.P.; Tetteh, K.K.; Moens, L.; Maizels, R.M. A novel C-type lectin secreted by a tissue-dwelling parasitic nematode. Curr. Biol. 1999, 9, 825–828. [Google Scholar] [CrossRef]
- Newton, S.E.; Monti, J.R.; Greenhalgh, C.J.; Ashman, K.; Meeusen, E.N. cDNA cloning of galectins from third stage larvae of the parasitic nematode Teladorsagia circumcincta. Mol. Biochem. Parasitol. 1997, 86, 143–153. [Google Scholar] [CrossRef]
- Harcus, Y.; Nicoll, G.; Murray, J.; Filbey, K.; Gomez-Escobar, N.; Maizels, R.M. C-type lectins from the nematode parasites Heligmosomoides polygyrus and Nippostrongylus brasiliensis. Parasitol. Int. 2009, 58, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Drickamer, K.; Dodd, R.B. C-Type lectin-like domains in Caenorhabditis elegans: Predictions from the complete genome sequence. Glycobiology 1999, 9, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Schulenburg, H.; Hoeppner, M.P.; Weiner, J.; Bornberg-Bauer, E. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology 2008, 213, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Pees, B.; Yang, W.; Zárate-Potes, A.; Schulenburg, H. Dierking, K. High innate immune specificity through diversified C-type lectin-like domain proteins in invertebrates. J. Innate. Immun. 2016, 8, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Wada, I.; Hasegawa, K.; Yorihuzi, T.; Tremblay, L.O.; Herscovics, A.; Nagata, K. A novel ER α-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001, 2, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Hirao, K.; Natsuka, Y.; Tamura, T.; Wada, I.; Morito, D.; Natsuka, S.; Romero, P.; Sleno, B.; Tremblay, L.O.; Herscovics, A.; et al. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J. Biol. Chem. 2006, 281, 9650–9658. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Angata, T. Siglecs—The major subfamily of I-type lectins. Glycobiology 2006, 16, 1R–27R. [Google Scholar] [CrossRef] [PubMed]
- Drickamer, K.; Fadden, A.J. Genomic analysis of C-type lectins. Biochem. Soc. Symp. 2002, 69, 59–72. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, C.; Zhang, Y.; Li, K.; Xu, L.; Guo, L.; Kong, Y.; Feng, L. Characterization and comparative analyses of two amphioxus intelectins involved in the innate immune response. Fish Shellfish Immunol. 2013, 34, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Xu, L.; Zhang, Y.; Zhang, C.; Zhang, C.; Zhao, F.; Feng, L. Comparative genomic and phylogenetic analyses of the intelectin gene family: Implications for their origin and evolution. Dev. Comp. Immunol. 2013, 41, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Bishnoi, R.; Khatri, I.; Subramanian, S.; Ramya, T.N. Prevalence of the F-type lectin domain. Glycobiology 2015, 25, 888–901. [Google Scholar] [CrossRef] [PubMed]
- Vasta, G.R.; Odom, E.W.; Bianchet, M.A.; Amzel, L.M.; Saito, K.; Ahmed, H. F-type lectins: A new family of recognition factors. In Animal Lectins: A Functional View; Vasta, G.R., Ahmed, H., Eds.; CRC Press: Boca Raton, FL, USA, 2008; pp. 465–474. [Google Scholar]
- Farrand, S.; Hotze, E.; Friese, P.; Hollingshead, S.K.; Smith, D.F.; Cummings, R.D.; Dale, G.L.; Tweten, R.K. Characterization of a streptococcal cholesterol-dependent cytolysin with a lewis Y and B specific lectin domain. Biochemistry 2008, 47, 7097–7107. [Google Scholar] [CrossRef] [PubMed]
- Leach, M.R.; Williams, D.B. Lectin-deficient calnexin is capable of binding class I histocompatibility molecules in vivo and preventing their degradation. J. Biol. Chem. 2004, 279, 9072–9079. [Google Scholar] [CrossRef] [PubMed]
- Thomson, S.P.; Williams, D.B. Delineation of the lectin site of the molecular chaperone calreticulin. Cell Stress Chaperones 2005, 10, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Grandhi, N.J.; Mamidi, A.S.; Surolia, A. Pattern recognition in legume lectins to extrapolate amino acid variability to sugar specificity. Adv. Exp. Med. Biol. 2015, 842, 199–215. [Google Scholar] [PubMed]
- Sharma, V.; Surolia, A. Analyses of carbohydrate recognition by legume lectins: Size of the combining site loops and their primary specificity. J. Mol. Biol. 1997, 267, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Itin, C.; Roche, A.C.; Monsigny, M.; Hauri, H.P. ERGIC-53 is a functional mannose-selective and calcium-dependent human homologue of leguminous lectins. Mol. Biol. Cell 1996, 7, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, Y.; Yamaguchi, Y.; Takahashi, N.; Arata, Y.; Kasai, K.; Ihara, Y.; Matsuo, I.; Ito, Y.; Yamamoto, K.; Kato, K. Sugar-binding properties of VIP36, an intracellular animal lectin operating as a cargo receptor. J. Biol. Chem. 2005, 280, 37178–37182. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Sobhany, M.; Transue, T.R.; Oguma, K.; Pedersen, L.C.; Negishi, M. Structural analysis by X-ray crystallography and calorimetry of a haemagglutinin component (HA1) of the progenitor toxin from Clostridium botulinum. Microbiology 2003, 149, 3361–3370. [Google Scholar] [CrossRef] [PubMed]
- Tenno, M.; Saeki, A.; Kezdy, F.J.; Elhammer, A.P.; Kurosaka, A. The lectin domain of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 1 is involved in O-glycosylation of a polypeptide with multiple acceptor sites. J. Biol. Chem. 2002, 277, 47088–47096. [Google Scholar] [CrossRef] [PubMed]
- Tateno, H.; Goldstein, I.J. Partial identification of carbohydrate-binding sites of a Galα1,3Galβ1,4GlcNAc-specific lectin from the mushroom Marasmius oreades by site-directed mutagenesis. Arch. Biochem. Biophys. 2004, 427, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T.A.; Revoredo, L.; Thome, J.J.; Tabak, L.A.; Vester-Christensen, M.B.; Clausen, H.; Gahlay, G.K.; Jarvis, D.L.; Johnson, R.W.; Moniz, H.A.; et al. The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J. Biol. Chem. 2013, 288, 19900–19914. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, T.; Onaga, S.; Murata, K.; Taira, T.; Katoh, E. LysM domains from Pteris ryukyuensis chitinase-A: A stability study and characterization of the chitin-binding site. J. Biol. Chem. 2008, 283, 5178–5187. [Google Scholar] [CrossRef] [PubMed]
- Mesnage, S.; Dellarole, M.; Baxter, N.J.; Rouget, J.B.; Dimitrov, J.D.; Wang, N.; Fujimoto, Y.; Hounslow, A.M.; Lacroix-Desmazes, S.; Fukase, K.; et al. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat. Commun. 2014, 5, 4269. [Google Scholar] [CrossRef] [PubMed]
- Sulzenbacher, G.; Roig-Zamboni, V.; Peumans, W.J.; Henrissat, B.; van Damme, E.J.; Bourne, Y. Structural basis for carbohydrate binding properties of a plant chitinase-like agglutinin with conserved catalytic machinery. J. Struct. Biol. 2015, 190, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Nemoto-Sasaki, Y.; Hayama, K.; Ohya, H.; Arata, Y.; Kaneko, M.K.; Saitou, N.; Hirabayashi, J.; Kasai, K. Caenorhabditis elegans galectins LEC-1-LEC-11: Structural features and sugar-binding properties. Biochim. Biophys. Acta 2008, 1780, 1131–1142. [Google Scholar] [CrossRef] [PubMed]
- Weis, W.I.; Drickamer, K.; Hendrickson, W.A. Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature 1992, 360, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Drickamer, K. Engineering galactose-binding activity into a C-type mannose-binding protein. Nature 1992, 360, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Childs, R.A.; Wright, J.R.; Ross, G.F.; Yuen, C.T.; Lawson, A.M.; Chai, W.; Drickamer, K.; Feizi, T. Specificity of lung surfactant protein SP-A for both the carbohydrate and the lipid moieties of certain neutral glycolipids. J. Biol. Chem. 1992, 267, 9972–9979. [Google Scholar] [PubMed]
- Nagae, M.; Ikeda, A.; Hanashima, S.; Kojima, T.; Matsumoto, N.; Yamamoto, K.; Yamaguchi, Y. Crystal structure of human dendritic cell inhibitory receptor C-type lectin domain reveals the binding mode with N-glycan. FEBS Lett. 2016, 590, 1280–1288. [Google Scholar] [CrossRef] [PubMed]
- Wood-Charlson, E.M.; Weis, V.M. The diversity of C-type lectins in the genome of a basal metazoan, Nematostella vectensis. Dev. Comp. Immunol. 2009, 33, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Gidrol, X.; Chrestin, H.; Tan, H.L.; Kush, A. Hevein, a lectin-like protein from Hevea brasiliensis (rubber tree) is involved in the coagulation of latex. J. Biol. Chem. 1994, 269, 9278–9283. [Google Scholar] [PubMed]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial peptides from plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef] [PubMed]
- Van Megen, H.; van den Elsen, S.; Holterman, M.; Karssen, G.; Mooyman, P.; Bongers, T.; Holovachov, O.; Bakker, J.; Helder, J. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 2009, 11, 927–950. [Google Scholar] [CrossRef]
- Kini, S.G.; Nguyen, P.Q.; Weissbach, S.; Mallagaray, A.; Shin, J.; Yoon, H.S.; Tam, J.P. Studies on the chitin binding property of novel cysteine-rich peptides from Alternanthera sessilis. Biochemistry 2015, 54, 6639–6649. [Google Scholar] [CrossRef] [PubMed]
- Asensio, J.L.; Canada, F.J.; Siebert, H.C.; Laynez, J.; Poveda, A.; Nieto, P.M.; Soedjanaamadja, U.M.; Gabius, H.J.; Jimenez-Barbero, J. Structural basis for chitin recognition by defense proteins: GlcNAc residues are bound in a multivalent fashion by extended binding sites in hevein domains. Chem. Biol. 2000, 7, 529–543. [Google Scholar] [CrossRef]
- Foote, H.C.; Ride, J.P.; Franklin-Tong, V.E.; Walker, E.A.; Lawrence, M.J.; Franklin, F.C. Cloning and expression of a distinctive class of self-incompatibility (S) gene from Papaver rhoeas L. Proc. Natl. Acad. Sci. USA 1994, 91, 2265–2269. [Google Scholar] [CrossRef] [PubMed]
- Zelensky, A.N.; Gready, J.E. Comparative analysis of structural properties of the C-type-lectin-like domain (CTLD). Proteins 2003, 52, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Tordai, H.; Banyai, L.; Patthy, L. The PAN module: The N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999, 461, 63–67. [Google Scholar] [CrossRef]
- Shen, Z.; Jacobs-Lorena, M. A type I peritrophic matrix protein from the malaria vector Anopheles gambiae binds to chitin. J. Biol. Chem. 1998, 273, 17665–17670. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Hofmann, K. The protease-associated domain: A homology domain associated with multiple classes of proteases. Trends Biochem. Sci. 2001, 26, 147–148. [Google Scholar] [CrossRef]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O.P. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.L.; Kaplan, D.T.; Dickson, D.W.; Mitchell, D.J. Root tissue response of two related soybean cultivars to infection by lectin-treated Meloidogyne spp. J. Nematol. 1989, 21, 219–228. [Google Scholar] [PubMed]
- McClure, M.A.; Stynes, B.A. Lectin binding sites on the amphidial exudates of Meloidogyne. J. Nematol. 1988, 20, 321–326. [Google Scholar] [PubMed]
- Spiegel, Y.; Cohn, E. Lectin binding to Meloidogyne javanica eggs. J. Nematol. 1982, 14, 406–407. [Google Scholar] [PubMed]
- Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 1998, 282, 2012–2018. [Google Scholar]
- Takeuchi, T.; Sennari, R.; Sugiura, K.; Tateno, H.; Hirabayashi, J.; Kasai, K. A C-type lectin of Caenorhabditis elegans: Its sugar-binding property revealed by glycoconjugate microarray analysis. Biochem. Biophys. Res. Commun. 2008, 377, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Arata, Y.; Hirabayashi, J.; Kasai, K. The two lectin domains of the tandem-repeat 32-kDa galectin of the nematode Caenorhabditis elegans have different binding properties. Studies with recombinant protein. J. Biochem. 1997, 121, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
- Jaouannet, M.; Magliano, M.; Arguel, M.J.; Gourgues, M.; Evangelisti, E.; Abad, P.; Rosso, M.N. The root-knot nematode calreticulin Mi-CRT is a key effector in plant defense suppression. Mol. Plant Microbe Interact. 2013, 26, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Mallo, G.V.; Kurz, C.L.; Couillault, C.; Pujol, N.; Granjeaud, S.; Kohara, Y.; Ewbank, J.J. Inducible antibacterial defense system in C. elegans. Curr. Biol. 2002, 12, 1209–1214. [Google Scholar] [CrossRef]
- Ideo, H.; Fukushima, K.; Gengyo-Ando, K.; Mitani, S.; Dejima, K.; Nomura, K.; Yamashita, K. A Caenorhabditis elegans glycolipid-binding galectin functions in host defense against bacterial infection. J. Biol. Chem. 2009, 284, 26493–26501. [Google Scholar] [CrossRef] [PubMed]
- Loukas, A.; Doedens, A.; Hintz, M.; Maizels, R.M. Identification of a new C-type lectin, TES-70, secreted by infective larvae of Toxocara canis, which binds to host ligands. Parasitology 2000, 121, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Ganji, S.; Jenkins, J.N.; Wubben, M.J. Molecular characterization of the reniform nematode C-type lectin gene family reveals a likely role in mitigating environmental stresses during plant parasitism. Gene 2014, 537, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, S.D.; Babayan, S.A.; Lhermitte-Vallarino, N.; Gray, N.; Xia, D.; Martin, C.; Kumar, S.; Taylor, D.W.; Blaxter, M.L.; Wastling, J.M.; et al. Comparative analysis of the secretome from a model filarial nematode (Litomosoides sigmodontis) reveals maximal diversity in gravid female parasites. Mol. Cell. Proteom. 2014, 13, 2527–2544. [Google Scholar] [CrossRef] [PubMed]
- Hewitson, J.P.; Harcus, Y.M.; Curwen, R.S.; Dowle, A.A.; Atmadja, A.K.; Ashton, P.D.; Wilson, A.; Maizels, R.M. The secretome of the filarial parasite, Brugia malayi: Proteomic profile of adult excretory-secretory products. Mol. Biochem. Parasitol. 2008, 160, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Geary, J.; Satti, M.; Moreno, Y.; Madrill, N.; Whitten, D.; Headley, S.A.; Agnew, D.; Geary, T.; Mackenzie, C. First analysis of the secretome of the canine heartworm, Dirofilaria immitis. Parasit. Vectors. 2012, 5, 140. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Cho, M.K.; Choi, S.H.; Lee, K.H.; Ahn, S.C.; Kim, D.H.; Yu, H.S. Inhibition of dextran sulfate sodium (DSS)-induced intestinal inflammation via enhanced IL-10 and TGF-β production by galectin-9 homologues isolated from intestinal parasites. Mol. Biochem. Parasitol. 2010, 174, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Hewitson, J.P.; Grainger, J.R.; Maizels, R.M. Helminth immunoregulation: The role of parasite secreted proteins in modulating host immunity. Mol. Biochem. Parasitol. 2009, 167, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rzepecka, J.; Rausch, S.; Klotz, C.; Schnoller, C.; Kornprobst, T.; Hagen, J.; Ignatius, R.; Lucius, R.; Hartmann, S. Calreticulin from the intestinal nematode Heligmosomoides polygyrus is a Th2-skewing protein and interacts with murine scavenger receptor-A. Mol. Immunol. 2009, 46, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Kasper, G.; Brown, A.; Eberl, M.; Vallar, L.; Kieffer, N.; Berry, C.; Girdwood, K.; Eggleton, P.; Quinnell, R.; Pritchard, D.I. A calreticulin-like molecule from the human hookworm Necator americanus interacts with C1q and the cytoplasmic signalling domains of some integrins. Parasite Immunol. 2001, 23, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, K.; Xie, H.; Wang, Y.T.; Wang, D.W.; Xu, C.L.; Huang, X.; Wang, D.S. A nematode calreticulin, Rs-CRT, is a key effector in reproduction and pathogenicity of Radopholus similis. PLoS ONE 2015, 10, e0129351. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Wei, L.; Chen, H.; Zhou, Y. Calreticulin is required for responding to stress, foraging, and fertility in the white-tip nematode, Aphelenchoides besseyi. Exp. Parasitol. 2015, 155, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.; Culerrier, R.; Barre, A.; Alvarez, R.; Rouge, P.; Peumans, W.J. A novel family of lectins evolutionarily related to class V chitinases: An example of neofunctionalization in legumes. Plant Physiol. 2007, 144, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Mast, S.W.; Diekman, K.; Karaveg, K.; Davis, A.; Sifers, R.N.; Moremen, K.W. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology 2005, 15, 421–436. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.B. Beyond lectins: The calnexin/calreticulin chaperone system of the endoplasmic reticulum. J. Cell Sci. 2006, 119, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Ghenea, S. Endoplasmic reticulum associated with degradation of glycoproteins in Caenorhabditis elegans. Rom. J. Biochem. 2009, 46, 53–62. [Google Scholar]
- Yoshida, Y.; Tanaka, K. Lectin-like ERAD players in ER and cytosol. Biochim. Biophys. Acta 2010, 1800, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.J.; Ryoo, H.D. Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc. Natl. Acad. Sci. USA 2009, 106, 17043–17048. [Google Scholar] [CrossRef] [PubMed]
- Jakob, C.A.; Bodmer, D.; Spirig, U.; Battig, P.; Marcil, A.; Dignard, D.; Bergeron, J.J.; Thomas, D.Y.; Aebi, M. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2001, 2, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Van Parijs, J.; Broekaert, W.F.; Goldstein, I.J.; Peumans, W.J. Hevein: An antifungal protein from rubber-tree (Hevea brasiliensis) latex. Planta 1991, 183, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.V.; Vyas, A.A.; Vyas, K.A.; Liu, Y.S.; Chiang, C.M.; Chi, L.M.; Wu, W.G. Heparin and heparan sulfate bind to snake cardiotoxin. Sulfated oligosaccharides as a potential target for cardiotoxin action. J. Biol. Chem. 1997, 272, 1484–1492. [Google Scholar] [CrossRef] [PubMed]
- Drenth, J.; Low, B.W.; Richardson, J.S.; Wright, C.S. The toxin-agglutinin fold. A new group of small protein structures organized around a four-disulfide core. J. Biol. Chem. 1980, 255, 2652–2655. [Google Scholar] [PubMed]
- Asensio, J.L.; Siebert, H.C.; von der Lieth, C.W.; Laynez, J.; Bruix, M.; Soedjanaamadja, U.M.; Beintema, J.J.; Canada, F.J.; Gabius, H.J.; Jimenez-Barbero, J. NMR investigations of protein-carbohydrate interactions: Studies on the relevance of Trp/Tyr variations in lectin binding sites as deduced from titration microcalorimetry and NMR studies on hevein domains. Determination of the NMR structure of the complex between pseudohevein and N,N′,N′′-triacetylchitotriose. Proteins 2000, 40, 218–236. [Google Scholar] [PubMed]
- Shukurov, R.R.; Voblikova, V.D.; Nikonorova, A.K.; Komakhin, R.A.; Komakhina, V.V.; Egorov, T.A.; Grishin, E.V.; Babakov, A.V. Transformation of tobacco and Arabidopsis plants with Stellaria media genes encoding novel hevein-like peptides increases their resistance to fungal pathogens. Transgenic Res. 2012, 21, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Yang, F.; Zhang, S.; Li, J.; Chen, H.; Liu, Q.; Zheng, J.; Xi, J.; Yi, K. Expression of a hevein-like gene in transgenic Agave hybrid No. 11648 enhances tolerance against zebra stripe disease. Plant Cell Tissue Organ Cult. 2014, 119, 579–585. [Google Scholar] [CrossRef]
- Slavokhotova, A.A.; Naumann, T.A.; Price, N.P.; Rogozhin, E.A.; Andreev, Y.A.; Vassilevski, A.A.; Odintsova, T.I. Novel mode of action of plant defense peptides—Hevein-like antimicrobial peptides from wheat inhibit fungal metalloproteases. FEBS J. 2014, 281, 4754–4764. [Google Scholar] [CrossRef] [PubMed]
- Engelmann, I.; Griffon, A.; Tichit, L.; Montanana-Sanchis, F.; Wang, G.; Reinke, V.; Waterston, R.H.; Hillier, L.W.; Ewbank, J.J. A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS ONE 2011, 6, e19055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, D.; Bazopoulou, D.; Pujol, N.; Tavernarakis, N.; Ewbank, J.J. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007, 8, R194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treitz, C.; Cassidy, L.; Hockendorf, A.; Leippe, M.; Tholey, A. Quantitative proteome analysis of Caenorhabditis elegans upon exposure to nematicidal Bacillus thuringiensis. J. Proteom. 2015, 113, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Twumasi-Boateng, K.; Shapira, M. Dissociation of immune responses from pathogen colonization supports pattern recognition in C. elegans. PLoS ONE 2012, 7, e35400. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Rae, R.; Iatsenko, I.; Sommer, R.J. System wide analysis of the evolution of innate immunity in the nematode model species Caenorhabditis elegans and Pristionchus pacificus. PLoS ONE 2012, 7, e44255. [Google Scholar] [CrossRef] [PubMed]
- Pujol, N.; Zugasti, O.; Wong, D.; Couillault, C.; Kurz, C.L.; Schulenburg, H.; Ewbank, J.J. Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 2008, 4, e1000105. [Google Scholar] [CrossRef] [PubMed]
- Dorris, M.; Viney, M.E.; Blaxter, M.L. Molecular phylogenetic analysis of the genus Strongyloides and related nematodes. Int. J. Parasitol. 2002, 32, 1507–1517. [Google Scholar] [CrossRef]
- Burnell, A.M.; Stock, S.P. Heterorhabditis, Steinernema and their bacterial symbionts—Lethal pathogens of insects. Nematology 2000, 2, 31–42. [Google Scholar] [CrossRef]
- Natarajan, K.; Dimasi, N.; Wang, J.; Mariuzza, R.A.; Marqulies, D.H. Structure and function of natural killer cell receptors: Multiple molecular solutions to self, nonself discrimination. Annu. Rev. Immunol. 2002, 20, 853–885. [Google Scholar] [CrossRef] [PubMed]
- Sano, H.; Kuroki, Y.; Honma, T.; Ogasawara, Y.; Sohma, H.; Voelker, D.R.; Akino, T. Analysis of chimeric proteins identifies the regions in the carbohydrate recognition domains of rat lung collectins that are essential for interactions with phospholipids, glycolipids, and alveolartype II cells. J. Biol. Chem. 1998, 273, 4783–4789. [Google Scholar] [CrossRef] [PubMed]
- Weiss, I.M.; Kaufmann, S.; Mann, K.; Fritz, M. Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusc Haliotis laevegata. Biochem. Biophys. Res. Commun. 2000, 267, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Ghedin, E.; Wang, S.; Spiro, D.; Caler, E.; Zhao, Q.; Crabtree, J.; Allen, J.E.; Delcher, A.L.; Guiliano, D.B.; Miranda-Saavedra, D.; et al. Draft genome of the filarial nematode parasite Brugia malayi. Science 2007, 317, 1756–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jex, A.R.; Liu, S.; Li, B.; Young, N.D.; Hall, R.S.; Li, Y.; Yang, L.; Zeng, N.; Xu, X.; Xiong, Z.; et al. Ascaris suum draft genome. Nature 2011, 479, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Cotton, J.A.; Lilley, C.J.; Jones, L.M.; Kikuchi, T.; Reid, A.J.; Thorpe, P.; Tsai, I.J.; Beasley, H.; Blok, V.; Cock, P.J.; et al. The genome and life-stage specific transcriptomes of Globodera pallida elucidate key aspects of plant parasitism by a cyst nematode. Genome Biol. 2014, 15, R43. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J.; et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef] [PubMed]
- Opperman, C.H.; Bird, D.M.; Williamson, V.M.; Rokhsar, D.S.; Burke, M.; Cohn, J.; Cromer, J.; Diener, S.; Gajan, J.; Graham, S.; et al. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc. Natl. Acad. Sci. USA 2008, 105, 14802–14807. [Google Scholar] [CrossRef] [PubMed]
- Abad, P.; Gouzy, J.; Aury, J.M.; Castagnone-Sereno, P.; Danchin, E.G.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008, 26, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Bauters, L.; Haegeman, A.; Kyndt, T.; Gheysen, G. Analysis of the transcriptome of Hirschmanniella oryzae to explore potential survival strategies and host-nematode interactions. Mol. Plant Pathol. 2013, 15, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Haegeman, A.; Bauters, L.; Kyndt, T.; Rahman, M.M.; Gheysen, G. Identification of candidate effector genes in the transcriptome of the rice root knot nematode Meloidogyne graminicola. Mol. Plant Pathol. 2012, 14, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Gantasala, N.P.; Roychowdhury, T.; Thakur, P.K.; Banakar, P.; Shukla, R.N.; Jones, M.G.; Rao, U. De novo transcriptome sequencing and analysis of the cereal cyst nematode, Heterodera avenae. PLoS ONE 2014, 9, e96311. [Google Scholar] [CrossRef] [PubMed]
- Haegeman, A.; Joseph, S.; Gheysen, G. Analysis of the transcriptome of the root lesion nematode Pratylenchus coffeae generated by 454 sequencing technology. Mol. Biochem. Parasitol. 2011, 178, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Nicol, P.; Gill, R.; Fosu-Nyarko, J.; Jones, M.G. De novo analysis and functional classification of the transcriptome of the root lesion nematode, Pratylenchus thornei, after 454 GS FLX sequencing. Int. J. Parasitol. 2012, 42, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Bolt, B.J.; Cain, S.; Chan, J.; Chen, W.J.; Davis, P.; Done, J.; Down, T.; Gao, S.; Grove, C.; et al. WormBase 2016: Expanding to enable helminth genomic research. Nucleic Acids Res. 2016, 44, D774–D780. [Google Scholar] [CrossRef] [PubMed]
- Milne, I.; Lindner, D.; Bayer, M.; Husmeier, D.; McGuire, G.; Marshall, D.F.; Wright, F. TOPALi v2: A rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 2009, 25, 126–127. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]

| Species | #Seq | Class V Chitinase | Hevein | Legume | LysM | F-Type Lectin | Ricin-B | Calnexin/Calreticulin | M-Type Lectin | C-Type Lectin | Galectin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genomes | CE | 26150 | 34 (30) | 3 | 2 (2) | 8 (?) | 1 (0) | 14 (6) | 2 (2) | 3 | 252 (52) | 38 (18) |
| AS | 18542 | 8 (4) | 0 | 3 (2) | 2 (?) | 1 (0) | 9 (8) | 3 (2) | 5 | 36 (8) | 11 (6) | |
| BM | 17938 | 12 (7) | 0 | 2 (2) | 4 (?) | 0 | 11 (10) | 8 (5) | 4 | 18 (1) | 13 (10) | |
| BX | 18074 | 7 (1) | 0 | 2 (2) | 1 (?) | 0 | 11 (7) | 3 (1) | 4 | 15 (0) | 14 (4) | |
| GP | 16417 | 9 (6) | 0 | 3 (2) | 0 | 0 | 9 (6) | 3 (2) | 5 | 22 (4) | 38 (7) | |
| MI | 20359 | 2 (0) | 0 | 1 (1) | 3 (?) | 0 | 7 (5) | 4 (3) | 6 | 57 (8) | 70 (9) | |
| MH | 14421 | 4 (3) | 0 | 2 (2) | 4 (?) | 0 | 8 (5) | 1 (0) | 4 | 29 (2) | 53 (3) | |
| Transcriptomes | MG | 10973 | 3 | 0 | 0 | 0 | 0 | 1 | 4 | 2 | 48 | 13 |
| HO | 17061 | 5 | 0 | 2 | 2 | 0 | 2 | 5 | 2 | 33 | 28 | |
| HA | 10070 | 3 | 0 | 2 | 2 | 0 | 6 | 1 | 4 | 1 | 9 | |
| HG | 12262 | 4 | 0 | 0 | 1 | 0 | 0 | 5 | 0 | 4 | 9 | |
| PC | 11507 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 0 | 3 | 7 | |
| PT | 19533 | 0 | 0 | 0 | 1 | 0 | 4 | 2 | 1 | 0 | 5 | |
| # | 91 | 3 | 19 | 29 | 2 | 84 | 42 | 40 | 518 | 308 | ||
| Class | Description | CE | AS | BM | GP | MI | MH | BX |
|---|---|---|---|---|---|---|---|---|
| I | 1 CTLD | 141 | 24 | 15 | 17 | 27 | 17 | 9 |
| II | 2–3 CTLD | 46 | 0 | 0 | 1 | 24 | 10 | 1 |
| III | 1–3 CTLD + 1–3 CUB | 28 | 0 | 1 | 0 | 0 | 1 | 1 |
| IV | 1–2 CTLD + 1–2 CW | 16 | 0 | 0 | 0 | 0 | 0 | 0 |
| V | 1 CTLD + 1–2 VWA | 14 | 9 | 0 | 0 | 0 | 0 | 0 |
| VI | Complex structure | 7 | 2 | 1 | 4 | 6 | 1 | 4 |
| VII* | 4 CTLD | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Conserved Residue | % Residue |
|---|---|
| C1 | 93.17 |
| C2 | 68.45 |
| C3 | 70.30 |
| C4 | 77.49 |
| W | 70.66 |
| G | 90.59 |
| L | 42.80 |
| α1A | 80.07 |
| β1’L | 64.94 |
| Both C bridges | 48.71 |
| Only C1-C4 bridge | 24.35 |
| Only C2-C3 bridge | 11.99 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauters, L.; Naalden, D.; Gheysen, G. The Distribution of Lectins across the Phylum Nematoda: A Genome-Wide Search. Int. J. Mol. Sci. 2017, 18, 91. https://doi.org/10.3390/ijms18010091
Bauters L, Naalden D, Gheysen G. The Distribution of Lectins across the Phylum Nematoda: A Genome-Wide Search. International Journal of Molecular Sciences. 2017; 18(1):91. https://doi.org/10.3390/ijms18010091
Chicago/Turabian StyleBauters, Lander, Diana Naalden, and Godelieve Gheysen. 2017. "The Distribution of Lectins across the Phylum Nematoda: A Genome-Wide Search" International Journal of Molecular Sciences 18, no. 1: 91. https://doi.org/10.3390/ijms18010091






