Dysbindin-1 Involvement in the Etiology of Schizophrenia
Abstract
:1. Introduction
2. Expression of Dysbindin-1 in the Brain and Its Biological Functions
3. Interaction of Dysbindin-1 with Cellular Proteins
4. Association of Dysbindin-1 with Schizophrenia
5. Dysbindin-1 Mutation Links to Schizophrenia-Like Behaviors
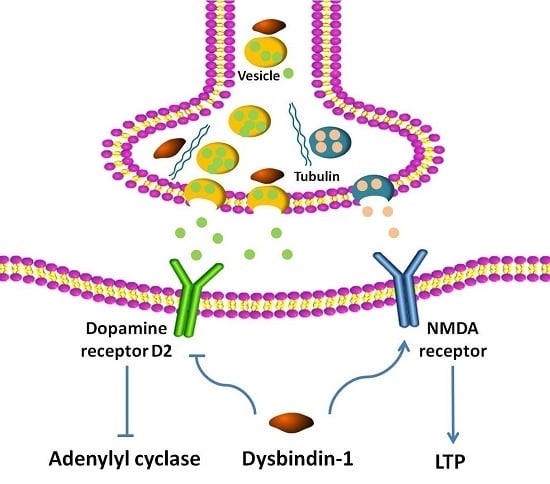
6. Regulation by Dysbindin-1 of Neurotransmitter Receptors
7. Dysbindin-1 Regulation of Neurite Outgrowth
8. Dysbindin-1 Is Required for Diverse Presynaptic and Postsynaptic Mechanisms
9. Summary, Conclusions and Perspective
Acknowledgments
Conflicts of Interest
References
- Wang, H.; Farhan, M.; Xu, J.; Lazarovici, P.; Zheng, W. The involvement of darpp-32 in the pathophysiology of schizophrenia. Oncotarget 2017, 8, 53791–53803. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Wang, H.; Zeng, Z.; Lin, J.; Little, P.J.; Srivastava, L.K.; Quirion, R. The possible role of the akt signaling pathway in schizophrenia. Brain Res. 2012, 1470, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Kantrowitz, J.T. Managing negative symptoms of schizophrenia: How far have we come? CNS Drugs 2017, 31, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Schooler, N.R.; Buchanan, R.W.; Laughren, T.; Leucht, S.; Nasrallah, H.A.; Potkin, S.G.; Abi-Saab, D.; Berardo, C.G.; Bugarski-Kirola, D.; Blaettler, T.; et al. Defining therapeutic benefit for people with schizophrenia: Focus on negative symptoms. Schizophr. Res. 2015, 162, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Snyder, G.L.; Vanover, K.E. Dopamine targeting drugs for the treatment of schizophrenia: Past, present and future. Curr. Top. Med. Chem. 2016, 16, 3385–3403. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Walton, N.M.; Yamada, H.; Kondo, Y.; Marek, G.J.; Tajinda, K. The impact of genetics on future drug discovery in schizophrenia. Expert. Opin. Drug Discov. 2017, 12, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Eyre, H.; Jacka, F.N.; Dodd, S.; Dean, O.; McEwen, S.; Debnath, M.; McGrath, J.; Maes, M.; Amminger, P.; et al. A review of vulnerability and risks for schizophrenia: Beyond the two hit hypothesis. Neurosci. Biobehav. Rev. 2016, 65, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Attademo, L.; Bernardini, F.; Garinella, R.; Compton, M.T. Environmental pollution and risk of psychotic disorders: A review of the science to date. Schizophr. Res. 2017, 181, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Wang, Y.; Li, Z.; Huang, J.; Lui, S.S.; Tan, S.P.; Yu, X.; Cheung, E.F.; He, M.G.; Ott, J.; et al. Heritability and familiality of neurological soft signs: Evidence from healthy twins, patients with schizophrenia and non-psychotic first-degree relatives. Psychol. Med. 2016, 46, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Owens, S.F.; Picchioni, M.M.; Rijsdijk, F.V.; Stahl, D.; Vassos, E.; Rodger, A.K.; Collier, D.A.; Murray, R.M.; Toulopoulou, T. Genetic overlap between episodic memory deficits and schizophrenia: Results from the maudsley twin study. Psychol. Med. 2011, 41, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Kendler, K.S.; Diehl, S.R. The genetics of schizophrenia: A current, genetic-epidemiologic perspective. Schizophr. Bull. 1993, 19, 261–285. [Google Scholar] [CrossRef] [PubMed]
- Riley, B.; Kendler, K.S. Molecular genetic studies of schizophrenia. Eur. J. Hum. Genet. 2006, 14, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Cardno, A.G.; Gottesman, I.I. Twin studies of schizophrenia: From bow-and-arrow concordances to star wars mx and functional genomics. Am. J. Med. Genet. 2000, 97, 12–17. [Google Scholar] [CrossRef]
- Tang, J.; Fan, Y.; Li, H.; Xiang, Q.; Zhang, D.F.; Li, Z.; He, Y.; Liao, Y.; Wang, Y.; He, F.; et al. Whole-genome sequencing of monozygotic twins discordant for schizophrenia indicates multiple genetic risk factors for schizophrenia. J. Genet. Genom. 2017, 44, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Owen, M.J.; Williams, N.M.; O’Donovan, M.C. Dysbindin-1 and schizophrenia: From genetics to neuropathology. J. Clin. Investig. 2004, 113, 1255–1257. [Google Scholar] [CrossRef] [PubMed]
- Prats, C.; Arias, B.; Moya-Higueras, J.; Pomarol-Clotet, E.; Parellada, M.; Gonzalez-Pinto, A.; Peralta, V.; Ibanez, M.I.; Martin, M.; Fananas, L.; et al. Evidence of an epistatic effect between dysbindin-1 and neuritin-1 genes on the risk for schizophrenia spectrum disorders. Eur. Psychiatry 2017, 40, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, G.; Morris, D.W.; Clarke, S.; McGhee, K.A.; Schwaiger, S.; Nangle, J.M.; Garavan, H.; Robertson, I.H.; Gill, M.; Corvin, A. Variance in neurocognitive performance is associated with dysbindin-1 in schizophrenia: A preliminary study. Neuropsychologia 2007, 45, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Markov, V.; Krug, A.; Krach, S.; Jansen, A.; Eggermann, T.; Zerres, K.; Stocker, T.; Shah, N.J.; Nothen, M.M.; Treutlein, J.; et al. Impact of schizophrenia-risk gene dysbindin 1 on brain activation in bilateral middle frontal gyrus during a working memory task in healthy individuals. Hum. Brain Mapp. 2010, 31, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Yang, F.; Xiao, Y.; Tan, S.; Husain, N.; Ren, M.; Hu, Z.; Martinowich, K.; Ng, J.S.; Kim, P.J.; et al. Regulation of brain-derived neurotrophic factor exocytosis and gamma-aminobutyric acidergic interneuron synapse by the schizophrenia susceptibility gene dysbindin-1. Biol. Psychiatry 2016, 80, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Shintani, N.; Onaka, Y.; Hashimoto, R.; Takamura, H.; Nagata, T.; Umeda-Yano, S.; Mouri, A.; Mamiya, T.; Haba, R.; Matsuzaki, S.; et al. Behavioral characterization of mice overexpressing human dysbindin-1. Mol. Brain 2014, 7, 74. [Google Scholar] [CrossRef] [PubMed]
- Petit, E.I.; Michalak, Z.; Cox, R.; O’Tuathaigh, C.M.; Clarke, N.; Tighe, O.; Talbot, K.; Blake, D.; Joel, J.; Shaw, A.; et al. Dysregulation of specialized delay/interference-dependent working memory following loss of dysbindin-1a in schizophrenia-related phenotypes. Neuropsychopharmacoloy 2017, 42, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Talbot, K.; Eidem, W.L.; Tinsley, C.L.; Benson, M.A.; Thompson, E.W.; Smith, R.J.; Hahn, C.G.; Siegel, S.J.; Trojanowski, J.Q.; Gur, R.E.; et al. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J. Clin. Investig. 2004, 113, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Weickert, C.S.; Straub, R.E.; McClintock, B.W.; Matsumoto, M.; Hashimoto, R.; Hyde, T.M.; Herman, M.M.; Weinberger, D.R.; Kleinman, J.E. Human dysbindin (DTNBP1) gene expression in normal brain and in schizophrenic prefrontal cortex and midbrain. Arch. Gen. Psychiatry 2004, 61, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Ghiani, C.A.; Starcevic, M.; Rodriguez-Fernandez, I.A.; Nazarian, R.; Cheli, V.T.; Chan, L.N.; Malvar, J.S.; de Vellis, J.; Sabatti, C.; Dell’Angelica, E.C. The dysbindin-containing complex (BLOC-1) in brain: Developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Mol. Psychiatry 2010, 15, 115. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Fei, E.; Fu, C.; Ren, H.; Wang, G. Dysbindin-1, a schizophrenia-related protein, facilitates neurite outgrowth by promoting the transcriptional activity of p53. Mol. Psychiatry 2011, 16, 1105–1116. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, F.; Burdick, M.C.; Callicott, J.H.; Weinberger, D.R. Epistatic interaction between comt and DTNBP1 modulates prefrontal function in mice and in humans. Mol. Psychiatry 2014, 19, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Yang, F.; Papaleo, F.; Wang, H.X.; Gao, W.J.; Weinberger, D.R.; Lu, B. Role of dysbindin in dopamine receptor trafficking and cortical gaba function. Proc. Natl. Acad. Sci. USA 2009, 106, 19593–19598. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; LeGros, R.P.; Louneva, N.; Yeh, L.; Cohen, J.W.; Hahn, C.G.; Blake, D.J.; Arnold, S.E.; Talbot, K. Dysbindin-1 in dorsolateral prefrontal cortex of schizophrenia cases is reduced in an isoform-specific manner unrelated to dysbindin-1 mrna expression. Hum. Mol. Genet. 2009, 18, 3851–3863. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yuan, Y.; Zhang, Z.; Yan, H.; Feng, Y.; Li, W. Dysbindin-1c is required for the survival of hilar mossy cells and the maturation of adult newborn neurons in dentate gyrus. J. Biol. Chem. 2014, 289, 29060–29072. [Google Scholar] [CrossRef] [PubMed]
- Straub, R.E.; Jiang, Y.; MacLean, C.J.; Ma, Y.; Webb, B.T.; Myakishev, M.V.; Harris-Kerr, C.; Wormley, B.; Sadek, H.; Kadambi, B.; et al. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am. J. Hum. Genet. 2002, 71, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Menke, A.; Jockusch, H. Decreased osmotic stability of dystrophin-less muscle cells from the mdx mouse. Nature 1991, 349, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Benson, M.A.; Newey, S.E.; Martin-Rendon, E.; Hawkes, R.; Blake, D.J. Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J. Biol. Chem. 2001, 276, 24232–24241. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Morishita, R.; Shinoda, T.; Iwamoto, I.; Sudo, K.; Okamoto, K.; Nagata, K. Dysbindin-1, wave2 and abi-1 form a complex that regulates dendritic spine formation. Mol. Psychiatry 2010, 15, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Nazarian, R.; Starcevic, M.; Spencer, M.J.; Dell’Angelica, E.C. Reinvestigation of the dysbindin subunit of bloc-1 (biogenesis of lysosome-related organelles complex-1) as a dystrobrevin-binding protein. Biochem. J. 2006, 395, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Falcon-Perez, J.M.; Starcevic, M.; Gautam, R.; Dell’Angelica, E.C. Bloc-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J. Biol. Chem. 2002, 277, 28191–28199. [Google Scholar] [CrossRef] [PubMed]
- Talbot, K.; Louneva, N.; Cohen, J.W.; Kazi, H.; Blake, D.J.; Arnold, S.E. Synaptic dysbindin-1 reductions in schizophrenia occur in an isoform-specific manner indicating their subsynaptic location. PLoS ONE 2011, 6, e16886. [Google Scholar] [CrossRef] [PubMed]
- Larimore, J.; Ryder, P.V.; Kim, K.Y.; Ambrose, L.A.; Chapleau, C.; Calfa, G.; Gross, C.; Bassell, G.J.; Pozzo-Miller, L.; Smith, Y.; et al. Mecp2 regulates the synaptic expression of a dysbindin-bloc-1 network component in mouse brain and human induced pluripotent stem cell-derived neurons. PLoS ONE 2013, 8, e65069. [Google Scholar] [CrossRef] [PubMed]
- Starcevic, M.; Dell’Angelica, E.C. Identification of snapin and three novel proteins (blos1, blos2, and blos3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (bloc-1). J. Biol Chem. 2004, 279, 28393–28401. [Google Scholar] [CrossRef] [PubMed]
- Dickman, D.K.; Davis, G.W. The schizophrenia susceptibility gene dysbindin controls synaptic homeostasis. Science 2009, 326, 1127–1130. [Google Scholar] [CrossRef] [PubMed]
- Newell-Litwa, K.; Salazar, G.; Smith, Y.; Faundez, V. Roles of bloc-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles. Mol. Biol. Cell 2009, 20, 1441–1453. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, Y.; Sei, Y.; Weinberger, D.R.; Straub, R.E. Evidence that the bloc-1 protein dysbindin modulates dopamine d2 receptor internalization and signaling but not d1 internalization. J. Neurosci. 2007, 27, 12390–12395. [Google Scholar] [CrossRef] [PubMed]
- Oyama, S.; Yamakawa, H.; Sasagawa, N.; Hosoi, Y.; Futai, E.; Ishiura, S. Dysbindin-1, a schizophrenia-related protein, functionally interacts with the DNA-dependent protein kinase complex in an isoform-dependent manner. PLoS ONE 2009, 4, e4199. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sun, Y.; Ye, H.; Zhu, L.; Liu, J.; Wu, X.; Wang, L.; He, T.; Shen, Y.; Wu, J.Y.; et al. Increased dysbindin-1b isoform expression in schizophrenia and its propensity in aggresome formation. Cell Discov. 2015, 1, 15032. [Google Scholar] [CrossRef] [PubMed]
- Hattori, S.; Murotani, T.; Matsuzaki, S.; Ishizuka, T.; Kumamoto, N.; Takeda, M.; Tohyama, M.; Yamatodani, A.; Kunugi, H.; Hashimoto, R. Behavioral abnormalities and dopamine reductions in sdy mutant mice with a deletion in DTNBP1, a susceptibility gene for schizophrenia. Biochem. Biophys. Res. Commun. 2008, 373, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.W.; Feng, Y.Q.; Hao, C.J.; Guo, X.L.; He, X.; Zhou, Z.Y.; Guo, N.; Huang, H.P.; Xiong, W.; Zheng, H.; et al. DTNBP1, a schizophrenia susceptibility gene, affects kinetics of transmitter release. J. Cell Biol. 2008, 181, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, F.; Yang, F.; Garcia, S.; Chen, J.; Lu, B.; Crawley, J.N.; Weinberger, D.R. Dysbindin-1 modulates prefrontal cortical activity and schizophrenia-like behaviors via dopamine/d2 pathways. Mol. Psychiatry 2012, 17, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Metzinger, L.; Blake, D.J.; Squier, M.V.; Anderson, L.V.; Deconinck, A.E.; Nawrotzki, R.; Hilton-Jones, D.; Davies, K.E. Dystrobrevin deficiency at the sarcolemma of patients with muscular dystrophy. Hum. Mol. Genet. 1997, 6, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Selemon, L.D.; Zecevic, N. Schizophrenia: A tale of two critical periods for prefrontal cortical development. Transl. Psychiatry 2015, 5, e623. [Google Scholar] [CrossRef] [PubMed]
- Soma, M.; Wang, M.; Suo, S.; Ishiura, S. Dysbindin-1, a schizophrenia-related protein, interacts with hdac3. Neurosci. Lett. 2014, 582, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Chen, D.; Chen, R.; Hu, Q.; Wang, G. The schizophrenia-related protein dysbindin-1a is degraded and facilitates nf-kappa b activity in the nucleus. PLoS ONE 2015, 10, e0132639. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.O.M. Dual constraints on synapse formation and regression in schizophrenia: Neuregulin, neuroligin, dysbindin, disc1, musk and agrin. Aust. N. Z. J. Psychiatry 2008, 42, 662–677. [Google Scholar] [CrossRef] [PubMed]
- Fei, E.; Ma, X.; Zhu, C.; Xue, T.; Yan, J.; Xu, Y.; Zhou, J.; Wang, G. Nucleocytoplasmic shuttling of dysbindin-1, a schizophrenia-related protein, regulates synapsin i expression. J. Biol. Chem. 2010, 285, 38630–38640. [Google Scholar] [CrossRef] [PubMed]
- Cha, D.S.; Kudlow, P.A.; Baskaran, A.; Mansur, R.B.; McIntyre, R.S. Implications of epigenetic modulation for novel treatment approaches in patients with schizophrenia. Neuropharmacology 2014, 77, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Boersma, M.C.; Dresselhaus, E.C.; De Biase, L.M.; Mihalas, A.B.; Bergles, D.E.; Meffert, M.K. A requirement for nuclear factor-kappab in developmental and plasticity-associated synaptogenesis. J. Neurosci. 2011, 31, 5414–5425. [Google Scholar] [CrossRef] [PubMed]
- Roussos, P.; Katsel, P.; Davis, K.L.; Giakoumaki, S.G.; Lencz, T.; Malhotra, A.K.; Siever, L.J.; Bitsios, P.; Haroutunian, V. Convergent findings for abnormalities of the nf-kappab signaling pathway in schizophrenia. Neuropsychopharmacology 2013, 38, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Sochocka, M.; Diniz, B.S.; Leszek, J. Inflammatory response in the CNS: Friend or foe? Mol. Neurobiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P.; Culmsee, C.; Yu, Z.; Camandola, S. Roles of nuclear factor kappab in neuronal survival and plasticity. J. Neurochem. 2000, 74, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Greenhill, S.D.; Juczewski, K.; de Haan, A.M.; Seaton, G.; Fox, K.; Hardingham, N.R. Adult cortical plasticity depends on an early postnatal critical period. Science 2015, 349, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Kim, S.M.; Suh, B.K.; Sun, H.Y.; Park, Y.U.; Hong, J.H.; Park, C.; Nguyen, M.D.; Nagata, K.; Yoo, J.Y.; et al. Disrupted-in-schizophrenia 1 (disc1) regulates dysbindin function by enhancing its stability. J. Biol. Chem. 2015, 290, 7087–7096. [Google Scholar] [CrossRef] [PubMed]
- Talbot, K.; Cho, D.S.; Ong, W.Y.; Benson, M.A.; Han, L.Y.; Kazi, H.A.; Kamins, J.; Hahn, C.G.; Blake, D.J.; Arnold, S.E. Dysbindin-1 is a synaptic and microtubular protein that binds brain snapin. Hum. Mol. Genet. 2006, 15, 3041–3054. [Google Scholar] [CrossRef] [PubMed]
- Ilardi, J.M.; Mochida, S.; Sheng, Z.H. Snapin: A snare-associated protein implicated in synaptic transmission. Nat. Neurosci. 1999, 2, 119–124. [Google Scholar] [PubMed]
- Feng, Y.Q.; Zhou, Z.Y.; He, X.; Wang, H.; Guo, X.L.; Hao, C.J.; Guo, Y.; Zhen, X.C.; Li, W. Dysbindin deficiency in sandy mice causes reduction of snapin and displays behaviors related to schizophrenia. Schizophr. Res. 2008, 106, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Farrell, M.S.; Werge, T.; Sklar, P.; Owen, M.J.; Ophoff, R.A.; O’Donovan, M.C.; Corvin, A.; Cichon, S.; Sullivan, P.F. Evaluating historical candidate genes for schizophrenia. Mol. Psychiatry 2015, 20, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Schwab, S.G.; Knapp, M.; Mondabon, S.; Hallmayer, J.; Borrmann-Hassenbach, M.; Albus, M.; Lerer, B.; Rietschel, M.; Trixler, M.; Maier, W.; et al. Support for association of schizophrenia with genetic variation in the 6p22.3 gene, dysbindin, in sib-pair families with linkage and in an additional sample of triad families. Am. J. Hum. Genet. 2003, 72, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Cheah, S.Y.; Lawford, B.R.; Young, R.M.; Morris, C.P.; Voisey, J. Dysbindin (DTNBP1) variants are associated with hallucinations in schizophrenia. Eur. Psychiatry J. Assoc. Eur. Psychiatr. 2015, 30, 486–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakanidze, G.; Brandl, E.J.; Hutzler, C.; Aurass, F.; Onken, S.; Rapp, M.A.; Puls, I. Association of dystrobrevin-binding protein 1 polymorphisms with sustained attention and set-shifting in schizophrenia patients. Neuropsychobiology 2016, 74, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.H.; Kim, J.S.; Ryu, S.; Oh, S.; Noh, J.; Lee, W.K.; Park, T.; Lee, Y.S.; Lee, D.; Kwon, J.S.; et al. Association of genetic variations in DTNBP1 with cognitive function in schizophrenia patients and healthy subjects. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012, 159B, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Trost, S.; Platz, B.; Usher, J.; Scherk, H.; Wobrock, T.; Ekawardhani, S.; Meyer, J.; Reith, W.; Falkai, P.; Gruber, O. The DTNBP1 (dysbindin-1) gene variant rs2619522 is associated with variation of hippocampal and prefrontal grey matter volumes in humans. Eur. Arch. Psychiatry Clin. Neurosci. 2013, 263, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Voisey, J.; Swagell, C.D.; Hughes, I.P.; Connor, J.P.; Lawford, B.R.; Young, R.M.; Morris, C.P. A polymorphism in the dysbindin gene (DTNBP1) associated with multiple psychiatric disorders including schizophrenia. Behav. Brain Funct. 2010, 6, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [Green Version]
- Cox, M.M.; Tucker, A.M.; Tang, J.; Talbot, K.; Richer, D.C.; Yeh, L.; Arnold, S.E. Neurobehavioral abnormalities in the dysbindin-1 mutant, sandy, on a c57bl/6j genetic background. Genes Brain Behav. 2009, 8, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Saggu, S.; Cannon, T.D.; Jentsch, J.D.; Lavin, A. Potential molecular mechanisms for decreased synaptic glutamate release in dysbindin-1 mutant mice. Schizophr. Res. 2013, 146, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, S.K.; Ryan, R.T.; Wong, T.P.; Srivastava, L.K. Loss of dysbindin-1, a risk gene for schizophrenia, leads to impaired group 1 metabotropic glutamate receptor function in mice. Front. Behav. Neurosci. 2015, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Papaleo, F.; Weinberger, D.R. Dysbindin and schizophrenia: It’s dopamine and glutamate all over again. Biol. Psychiatry 2011, 69, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Kendler, K.S. Schizophrenia genetics and dysbindin: A corner turned? Am. J. Psychiatry 2004, 161, 1533–1536. [Google Scholar] [CrossRef] [PubMed]
- Talbot, K. The sandy (sdy) mouse: A dysbindin-1 mutant relevant to schizophrenia research. Prog. Brain Res. 2009, 179, 87–94. [Google Scholar] [PubMed]
- Takao, K.; Toyama, K.; Nakanishi, K.; Hattori, S.; Takamura, H.; Takeda, M.; Miyakawa, T.; Hashimoto, R. Impaired long-term memory retention and working memory in sdy mutant mice with a deletion in DTNBP1, a susceptibility gene for schizophrenia. Mol. Brain 2008, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Coyle, J.T. Schizophrenia: Basic and clinical. Adv. Neurobiol. 2017, 15, 255–280. [Google Scholar] [PubMed]
- Carr, G.V.; Jenkins, K.A.; Weinberger, D.R.; Papaleo, F. Loss of dysbindin-1 in mice impairs reward-based operant learning by increasing impulsive and compulsive behavior. Behav. Brain Res. 2013, 241, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Braff, D.L.; Geyer, M.A.; Swerdlow, N.R. Human studies of prepulse inhibition of startle: Normal subjects, patient groups, and pharmacological studies. Psychopharmacology 2001, 156, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Karlsgodt, K.H.; Robleto, K.; Trantham-Davidson, H.; Jairl, C.; Cannon, T.D.; Lavin, A.; Jentsch, J.D. Reduced dysbindin expression mediates n-methyl-d-aspartate receptor hypofunction and impaired working memory performance. Biol. Psychiatry 2011, 69, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Abi-Dargham, A. A dual hit model for dopamine in schizophrenia. Biol. Psychiatry 2017, 81, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.J.; Chohan, M.O.; Slifstein, M.; Kegeles, L.S.; Moore, H.; Abi-Dargham, A. Pathway-specific dopamine abnormalities in schizophrenia. Biol. Psychiatry 2017, 81, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Horga, G.; Cassidy, C.M.; Xu, X.; Moore, H.; Slifstein, M.; Van Snellenberg, J.X.; Abi-Dargham, A. Dopamine-related disruption of functional topography of striatal connections in unmedicated patients with schizophrenia. JAMA Psychiatry 2016, 73, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Schmieg, N.; Rocchi, C.; Romeo, S.; Maggio, R.; Millan, M.J.; Mannoury la Cour, C. Dysbindin-1 modifies signaling and cellular localization of recombinant, human d(3) and d(2) receptors. J. Neurochem. 2016, 136, 1037–1051. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.T.; Yang, F.; Chen, B.S.; Lu, Y.; Ji, Y.; Roche, K.W.; Lu, B. Dysbindin regulates hippocampal ltp by controlling nmda receptor surface expression. Proc. Natl. Acad. Sci. USA 2009, 106, 21395–21400. [Google Scholar] [CrossRef] [PubMed]
- Glen, W.B., Jr.; Horowitz, B.; Carlson, G.C.; Cannon, T.D.; Talbot, K.; Jentsch, J.D.; Lavin, A. Dysbindin-1 loss compromises nmdar-dependent synaptic plasticity and contextual fear conditioning. Hippocampus 2014, 24, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Jentsch, J.D.; Trantham-Davidson, H.; Jairl, C.; Tinsley, M.; Cannon, T.D.; Lavin, A. Dysbindin modulates prefrontal cortical glutamatergic circuits and working memory function in mice. Neuropsychopharmacology 2009, 34, 2601–2608. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Kulhara, P. What is schizophrenia: A neurodevelopmental or neurodegenerative disorder or a combination of both? A critical analysis. Indian J. Psychiatry 2010, 52, 21–27. [Google Scholar] [PubMed]
- Lefebvre, J.L.; Sanes, J.R.; Kay, J.N. Development of dendritic form and function. Annu. Rev. Cell Dev. Biol. 2015, 31, 741–777. [Google Scholar] [CrossRef] [PubMed]
- Kubota, K.; Kumamoto, N.; Matsuzaki, S.; Hashimoto, R.; Hattori, T.; Okuda, H.; Takamura, H.; Takeda, M.; Katayama, T.; Tohyama, M. Dysbindin engages in c-jun n-terminal kinase activity and cytoskeletal organization. Biochem. Biophys. Res. Commun. 2009, 379, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, A.; Tomoda, T.; Chang, J.; Takaki, M.; Zhan, C.; Morita, M.; Cascio, M.B.; Elashvili, S.; Koizumi, H.; Takanezawa, Y.; et al. Disc1-ndel1/nudel protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of disc1. Hum. Mol. Genet. 2006, 15, 3313–3323. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yan, W.; Chen, X. P53 is required for nerve growth factor-mediated differentiation of pc12 cells via regulation of trka levels. Cell Death Differ. 2006, 13, 2118–2128. [Google Scholar] [CrossRef] [PubMed]
- Ryder, P.V.; Faundez, V. Schizophrenia: The “bloc” may be in the endosomes. Sci. Signal 2009, 2, pe66. [Google Scholar] [CrossRef] [PubMed]
- Mead, C.L.; Kuzyk, M.A.; Moradian, A.; Wilson, G.M.; Holt, R.A.; Morin, G.B. Cytosolic protein interactions of the schizophrenia susceptibility gene dysbindin. J. Neurochem. 2010, 113, 1491–1503. [Google Scholar] [CrossRef] [PubMed]
- Sudhof, T.C. The synaptic vesicle cycle. Annu. Rev. Neurosci. 2004, 27, 509–547. [Google Scholar] [CrossRef] [PubMed]
- Hikita, T.; Taya, S.; Fujino, Y.; Taneichi-Kuroda, S.; Ohta, K.; Tsuboi, D.; Shinoda, T.; Kuroda, K.; Funahashi, Y.; Uraguchi-Asaki, J.; et al. Proteomic analysis reveals novel binding partners of dysbindin, a schizophrenia-related protein. J. Neurochem. 2009, 110, 1567–1574. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, A.; Hartwig, C.; Freeman, A.H.; Das, R.; Zlatic, S.A.; Vistein, R.; Burch, A.; Carrot, G.; Lewis, A.F.; Nelms, S.; et al. The proteome of bloc-1 genetic defects identifies the arp2/3 actin polymerization complex to function downstream of the schizophrenia susceptibility factor dysbindin at the synapse. J. Neurosci. 2016, 36, 12393–12411. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.H.; Racz, B.; Wang, H.; Burianek, L.; Weinberg, R.; Yasuda, R.; Wetsel, W.C.; Soderling, S.H. Disruption of arp2/3 results in asymmetric structural plasticity of dendritic spines and progressive synaptic and behavioral abnormalities. J. Neurosci. 2013, 33, 6081–6092. [Google Scholar] [CrossRef] [PubMed]
- Rocca, D.L.; Amici, M.; Antoniou, A.; Blanco Suarez, E.; Halemani, N.; Murk, K.; McGarvey, J.; Jaafari, N.; Mellor, J.R.; Collingridge, G.L.; et al. The small gtpase arf1 modulates arp2/3-mediated actin polymerization via pick1 to regulate synaptic plasticity. Neuron 2013, 79, 293–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, J.M.; Hu, Z.; Nordman, J.; Li, Z. The schizophrenia susceptibility gene dysbindin regulates dendritic spine dynamics. J. Neurosci. 2014, 34, 13725–13736. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, A.; Mullin, A.P.; Zlatic, S.A.; Easley, C.A.; Merritt, M.E.; Raj, N.; Larimore, J.; Gordon, D.E.; Peden, A.A.; Sanyal, S.; et al. The n-ethylmaleimide-sensitive factor and dysbindin interact to modulate synaptic plasticity. J. Neurosci. 2015, 35, 7643–7653. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, A.; Larimore, J.; Werner, E.; So, L.; Moreno-De-Luca, A.; Lese-Martin, C.; Lupashin, V.V.; Smith, Y.; Faundez, V. Quantitative proteomic and genetic analyses of the schizophrenia susceptibility factor dysbindin identify novel roles of the biogenesis of lysosome-related organelles complex 1. J. Neurosci. 2012, 32, 3697–3711. [Google Scholar] [CrossRef] [PubMed]
- Han, M.H.; Hu, Z.; Chen, C.Y.; Chen, Y.; Gucek, M.; Li, Z.; Markey, S.P. Dysbindin-associated proteome in the p2 synaptosome fraction of mouse brain. J. Proteome Res. 2014, 13, 4567–4580. [Google Scholar] [CrossRef] [PubMed]
- Guillozet-Bongaarts, A.L.; Hyde, T.M.; Dalley, R.A.; Hawrylycz, M.J.; Henry, A.; Hof, P.R.; Hohmann, J.; Jones, A.R.; Kuan, C.L.; Royall, J.; et al. Altered gene expression in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol. Psychiatry 2014, 19, 478–485. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Xu, J.; Lazarovici, P.; Zheng, W. Dysbindin-1 Involvement in the Etiology of Schizophrenia. Int. J. Mol. Sci. 2017, 18, 2044. https://doi.org/10.3390/ijms18102044
Wang H, Xu J, Lazarovici P, Zheng W. Dysbindin-1 Involvement in the Etiology of Schizophrenia. International Journal of Molecular Sciences. 2017; 18(10):2044. https://doi.org/10.3390/ijms18102044
Chicago/Turabian StyleWang, Haitao, Jiangping Xu, Philip Lazarovici, and Wenhua Zheng. 2017. "Dysbindin-1 Involvement in the Etiology of Schizophrenia" International Journal of Molecular Sciences 18, no. 10: 2044. https://doi.org/10.3390/ijms18102044