E-Learning for Rare Diseases: An Example Using Fabry Disease
Abstract
:1. Introduction
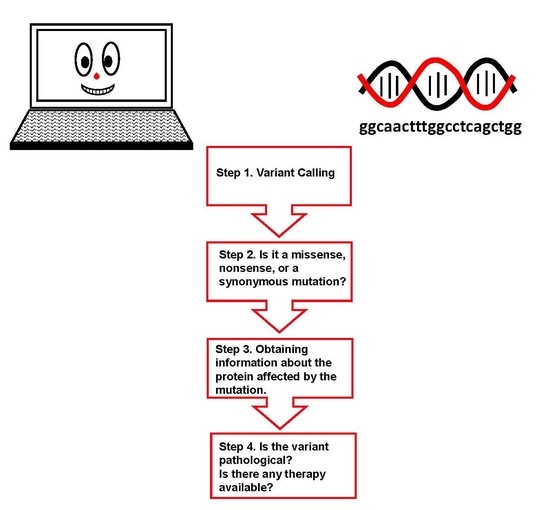
2. Results
- (1)
- Alpha-galactosidase A (AGAL) is encoded by the gene GLA (in the header of the file).
- (2)
- AGAL has catalytic activity: “Hydrolysis of terminal, non-reducing α-d-galactose residues in α-d-galactosides, including galactose oligosaccharides, galactomannans and galactolipids.” (in the “Function” section).
- (3)
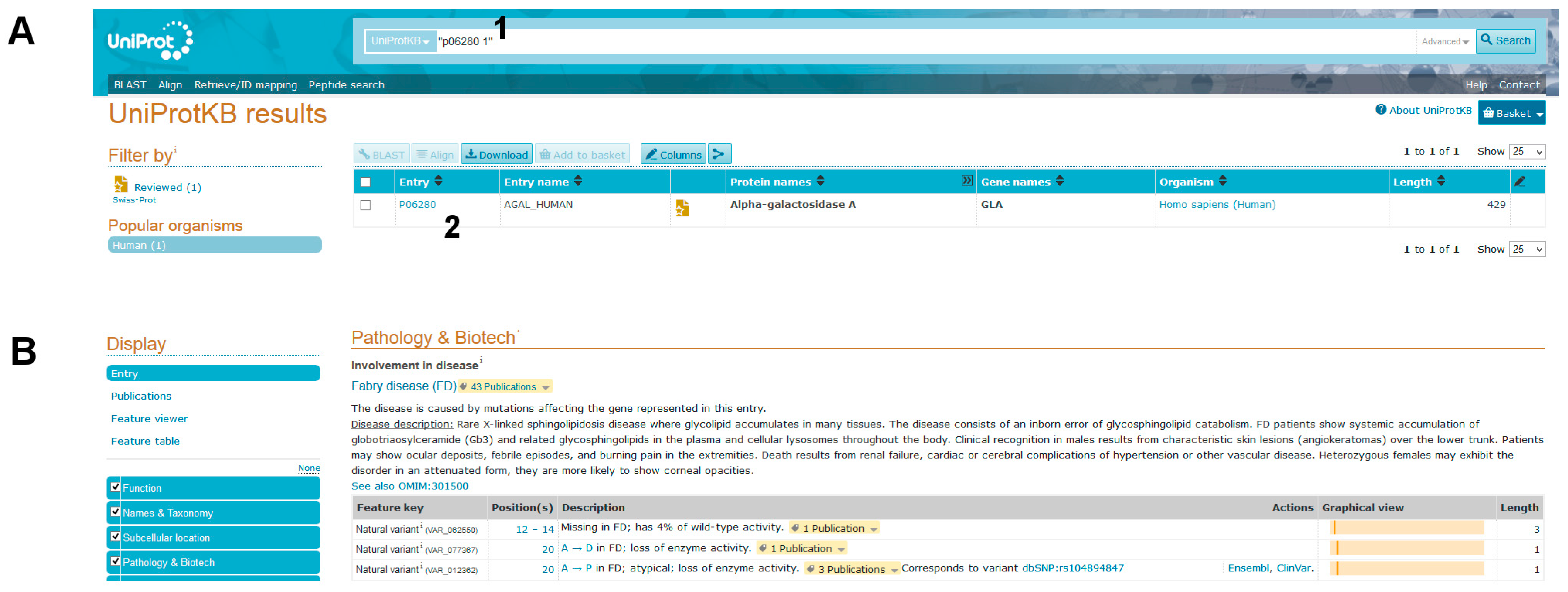
- AGAL is involved in a disease, Fabry disease (FD). A link to OMIM [2] (301500) provides clinical information about the disease (“Pathology & Biotech: Involvement in disease” section).
- (4)
- AGAL has a pharmaceutical use: “Available under the names Replagal® (from Shire) and Fabrazyme® (from Genzyme). Used as a long-term enzyme replacement therapy in patients with a confirmed diagnosis of Fabry disease. The differences between Replagal® (also known as agalsidase alpha) and Fabrazyme® (also known as agalsidase beta) lie in the glycosylation patterns. Agalsidase alpha is produced in the hamster CHO cell line while agalsidase alpha is produced in human cell lines.” (“Pathology & Biotech: Pharmaceutical use” section).
- (5)
- Another therapy is available for FD. A link to DrugBank (“Pathology & Biotech: Chemistry databases” section) [29] (DB05018) provides some details about the drug and summarizes its mechanism of action: “migalastat hydrochloride is an experimental, oral therapy for the treatment of Fabry disease and belongs to a class of molecules known as pharmacological chaperones”. Indeed, migalastat hydrochloride or 1-deoxygalactonojirimycin (DGJ) is a pharmacological chaperone for FD; it stabilizes wild type AGAL as well as some mutant forms. Mutations affecting the active site or cysteines involved in disulphide bridge formation do not respond. These conditions are necessary, but not sufficient to exclude the usefulness of migalastat [30]. In general, each mutation must be experimentally tested; the techniques needed for analysis have been extensively described elsewhere [31,32,33] but are outside the scope of this tutorial.
3. Discussion
4. Methods
4.1. Aims
4.2. Requirements
- UCSC Genome Browser, https://genome.ucsc.edu/
- UniProt, http://www.uniprot.org/
- PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/ [28]
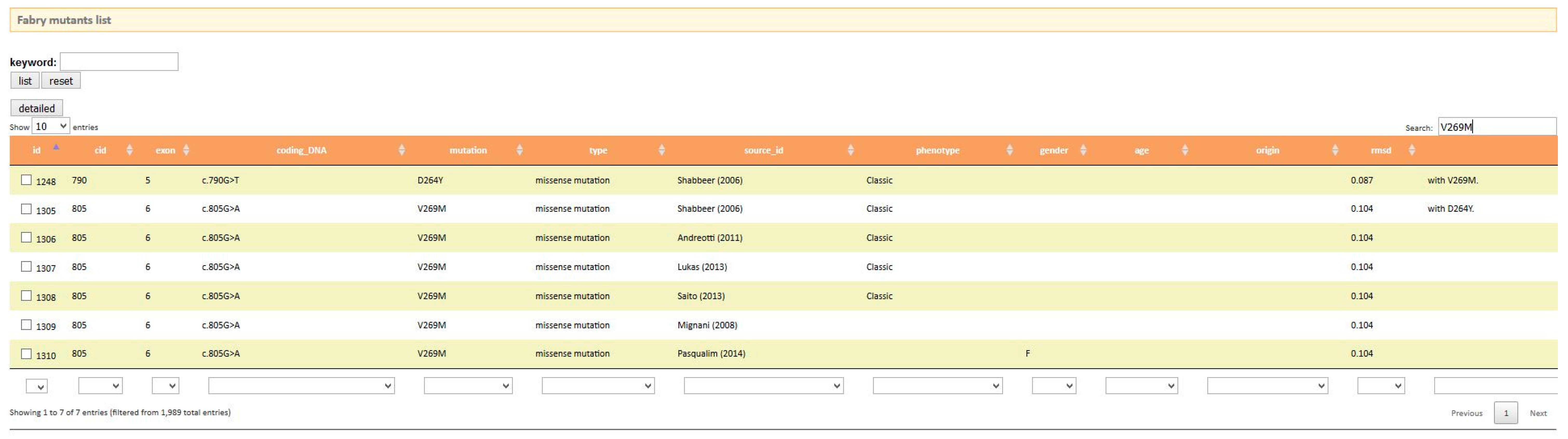
- fabry-database, http://fabry-database.org/ [49]
- Fabry_CEP, http://www1.na.icb.cnr.it/project/fabry_cep/ [39]. This tool can be run on-line or by locally downloading the supplementary material. Please download, unzip and press on index.html icon.
4.3. Input Exonic Sequence
4.4. Protocol
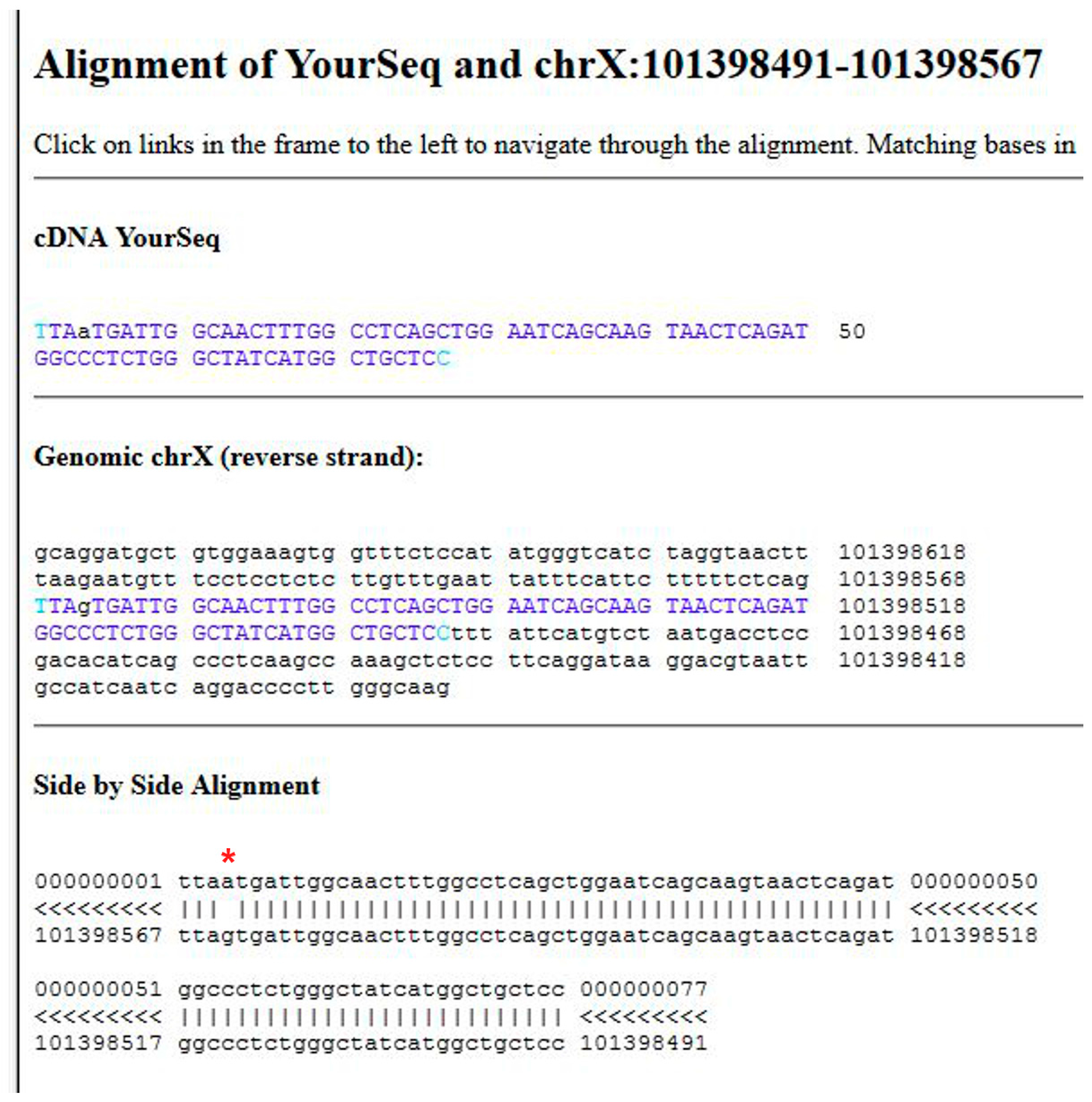
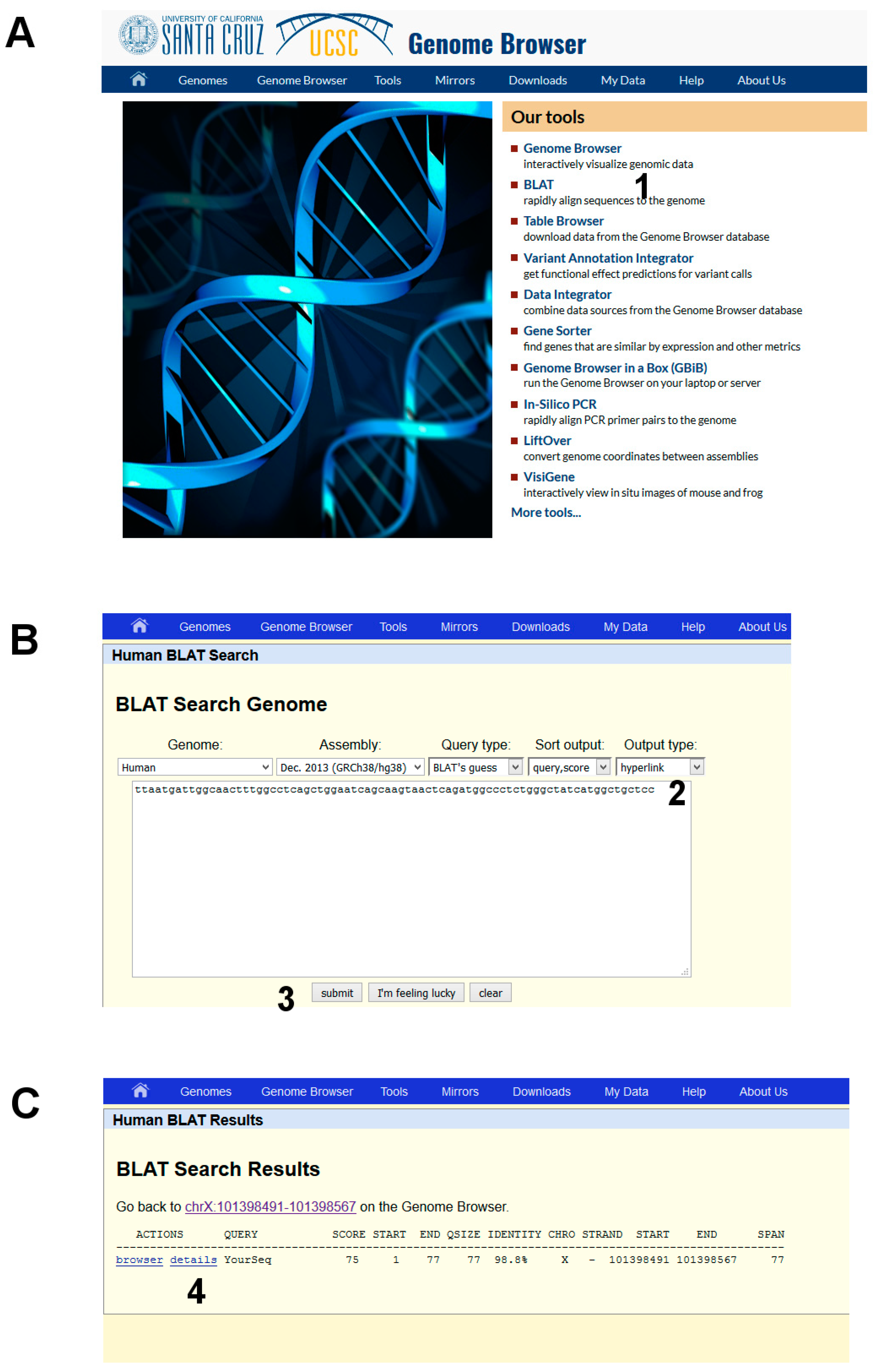
- (1)
- Open the UCSC genome browser and choose among the BLAT tools (Figure 6A, point 1).
- (2)
- The latest assembly of the HUMAN genome is chosen by default and does not need to be changed. An overview of how the program BLAT works is offered in the search page. Paste the given sequence into the Query Sequence box (Figure 6B, point 2).
- (3)
- Submit (Figure 6B, point 3).
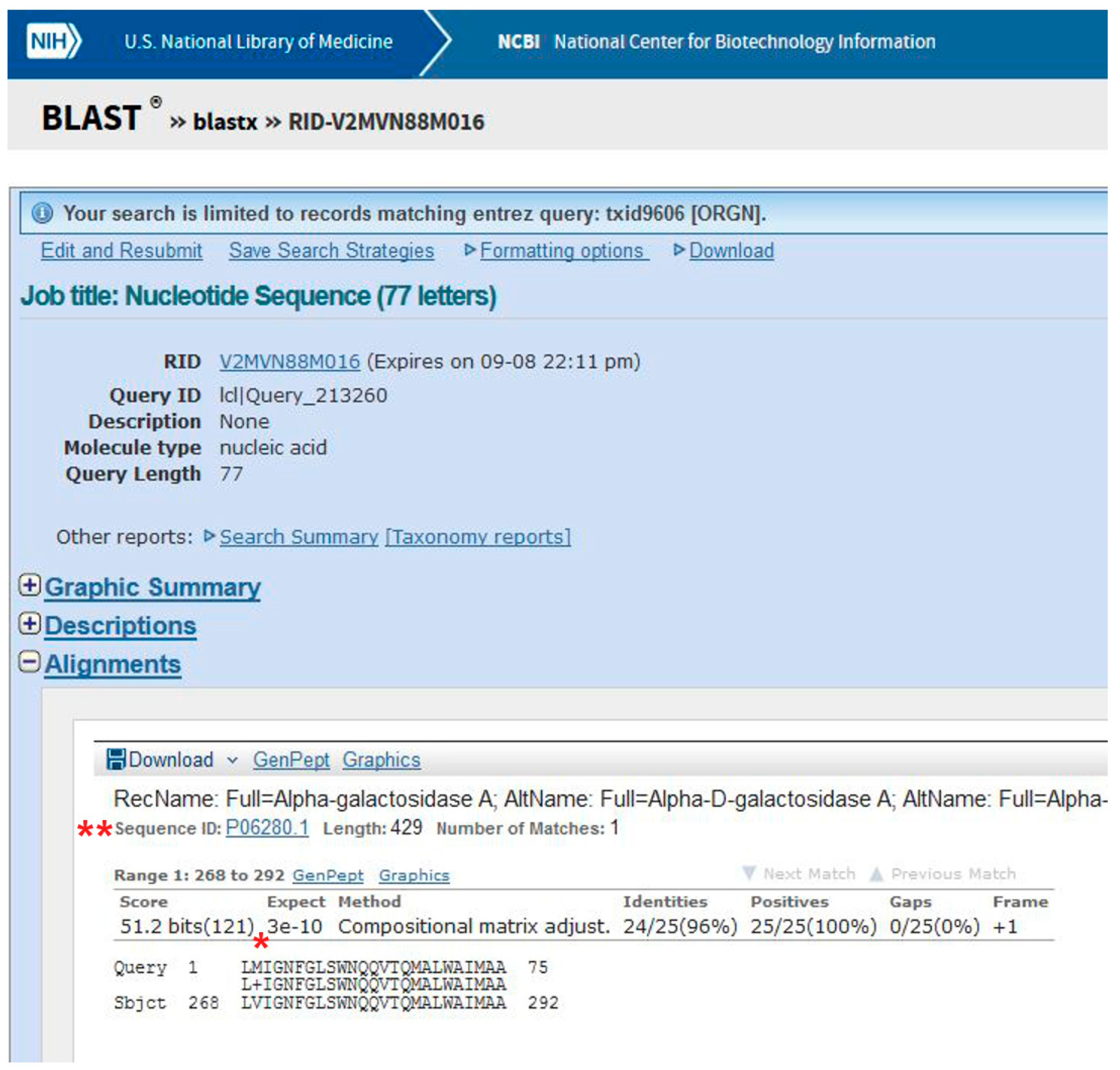
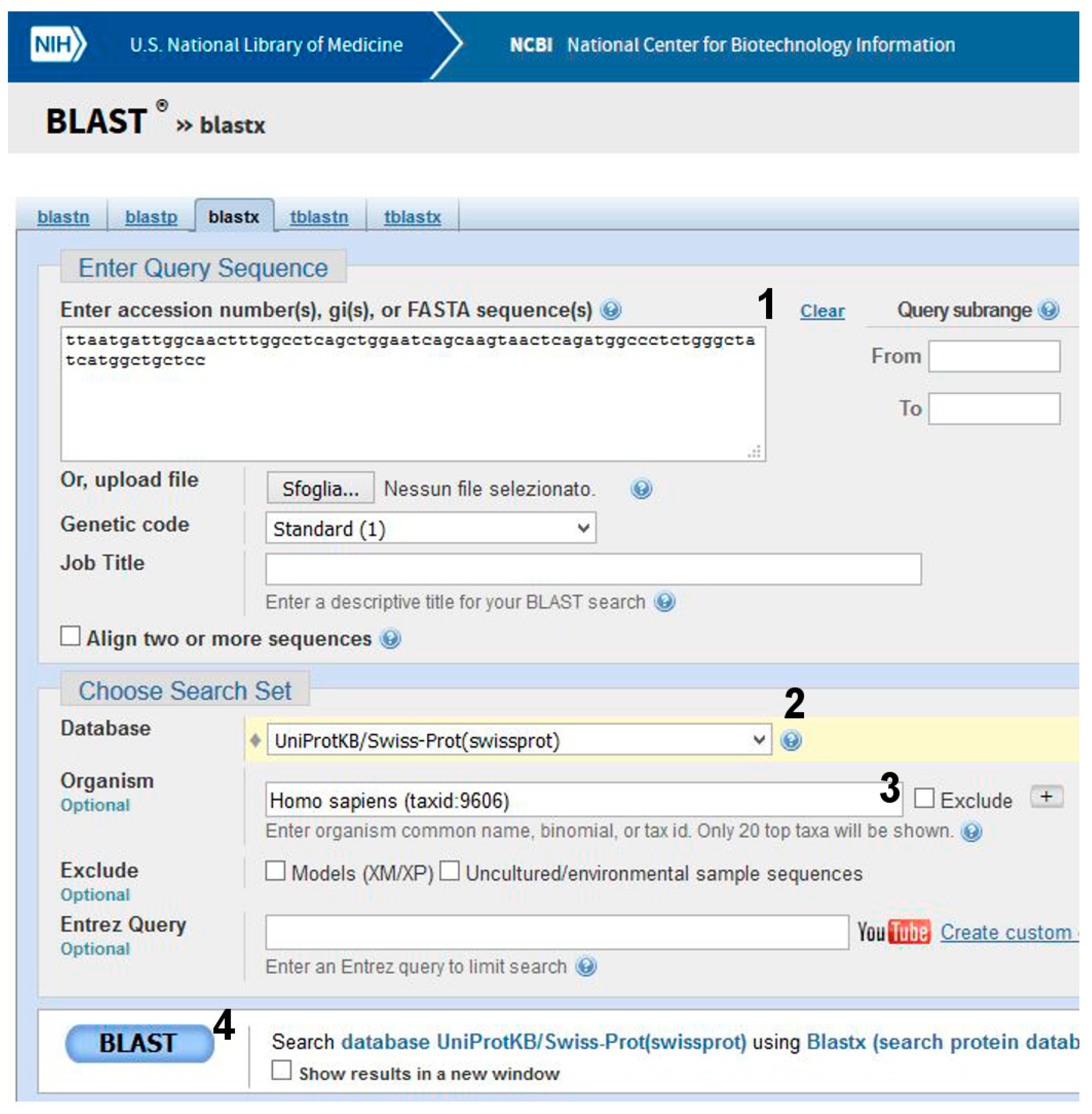
- (1)
- Go to BLAST and choose BlastX. The program searches protein databases using a translated nucleotide query.
- (2)
- (3)
- Click on BLAST (Figure 7, point 4).
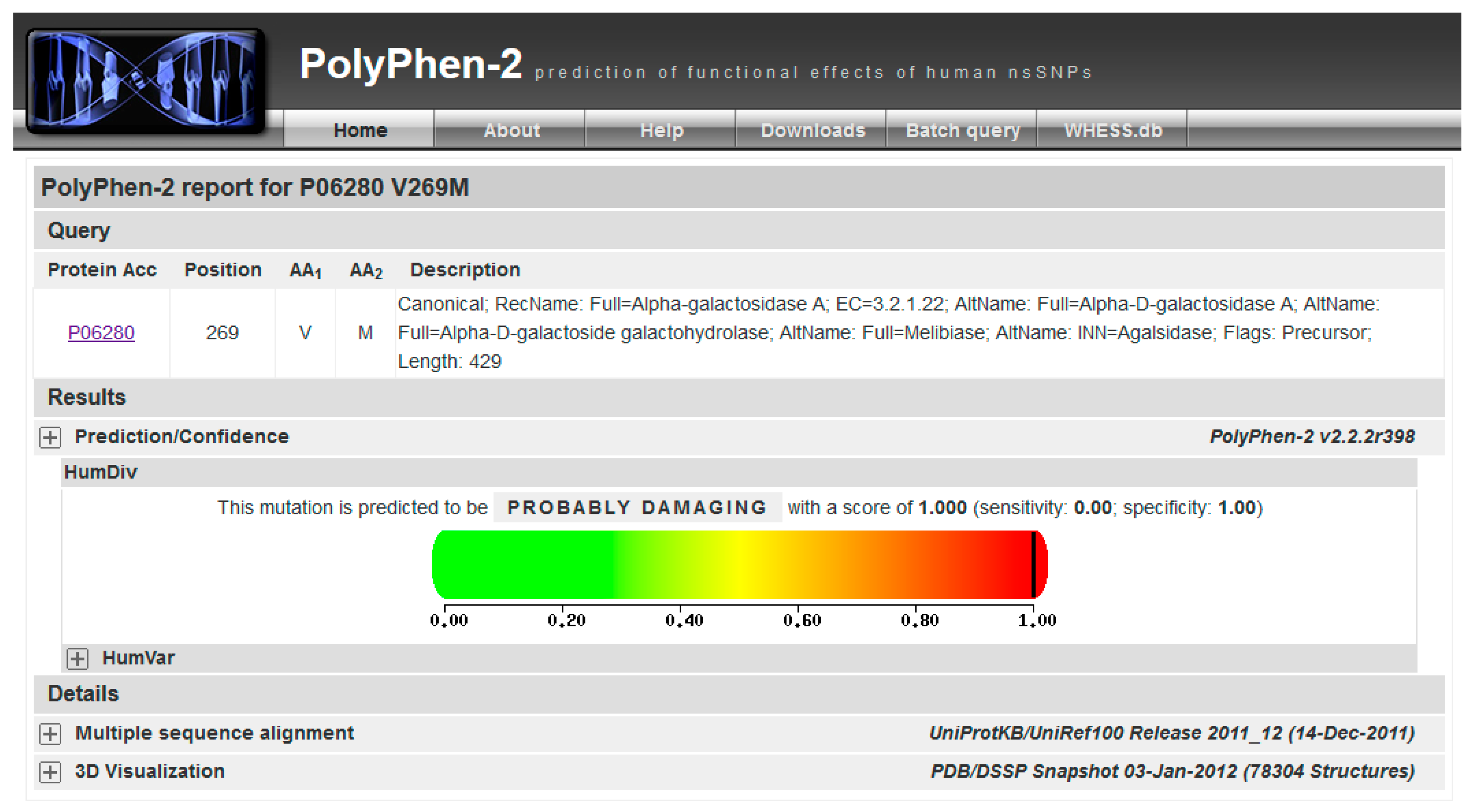
- (1)
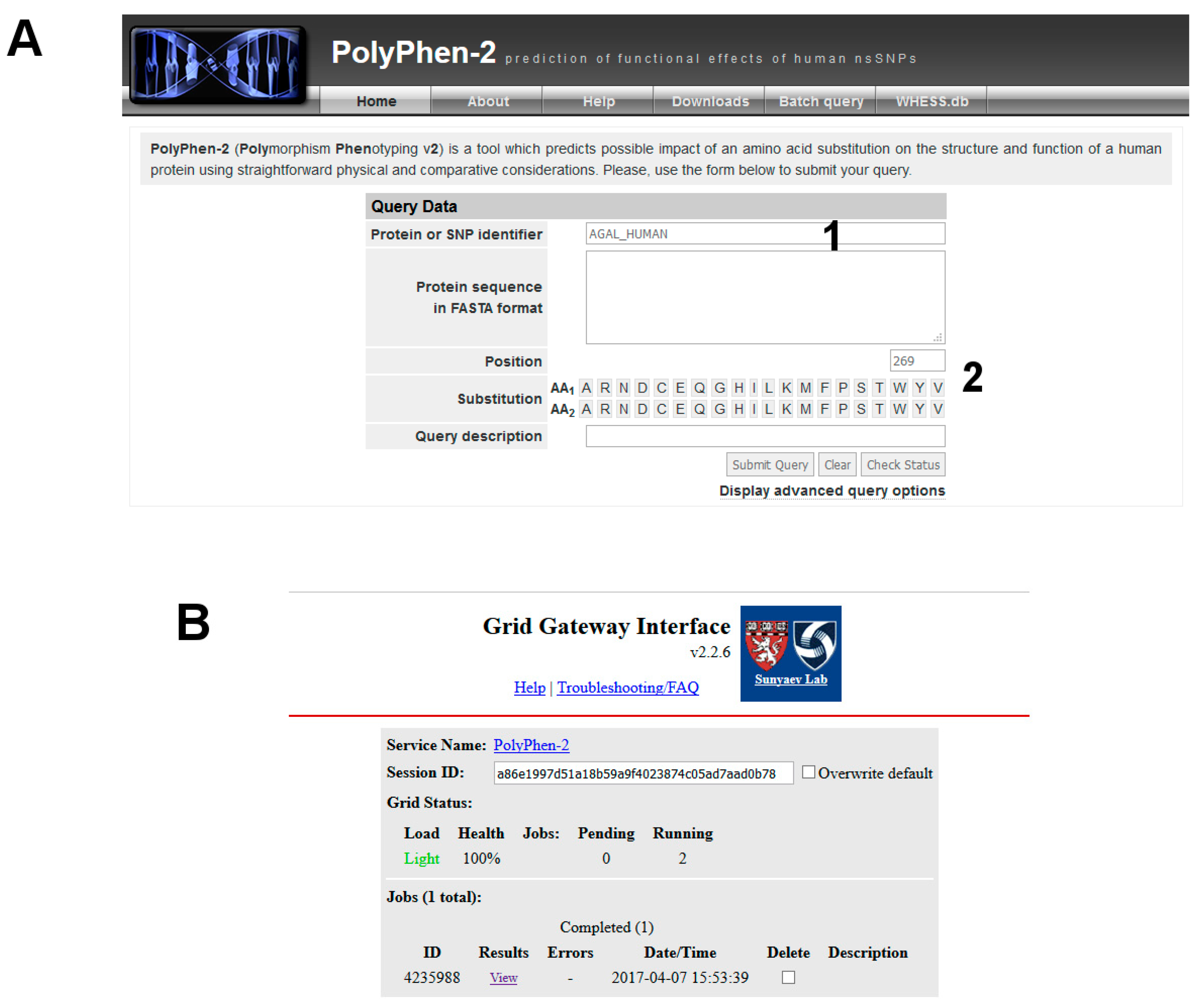
- Query Uniprot using the ID of the target protein found with BlastX: P06280.1 (Figure 8A, point1).
- (1)
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| FD | Fabry disease |
| Gb3 | Globotriaosylceramide |
| ERT | Enzymatic replacement therapy |
| DGJ | 1-Deoxygalactonojirimycin |
References
- Orphanet. Available online: http://www.Orpha.Net/consor/cgi-bin/index.Php (accessed on 19 September 2017).
- Online Mendelian Inheritance in Man, O. Mckusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). Available online: https://omim.Org/ (accessed on 19 September 2017).
- Germain, D.P. Fabry disease. Orphanet J. Rare Dis. 2010, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Linhart, A.; Elliott, P.M. The heart in anderson-fabry disease and other lysosomal storage disorders. Heart 2007, 93, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Schiffmann, R.; Moore, D.F. Neurological manifestations of fabry disease. In Fabry Disease: Perspectives from 5 Years of FOS; Oxford PharmaGenesis: Oxford, UK, 2006. [Google Scholar]
- Sunder-Plassmann, G. Renal manifestations of fabry disease. In Fabry Disease: Perspectives from 5 Years of FOS; Oxford PharmaGenesis: Oxford, UK, 2006. [Google Scholar]
- Citro, V.; Cammisa, M.; Liguori, L.; Cimmaruta, C.; Lukas, J.; Cubellis, M.V.; Andreotti, G. The large phenotypic spectrum of fabry disease requires graduated diagnosis and personalized therapy: A meta-analysis can help to differentiate missense mutations. Int. J. Mol. Sci. 2016, 17, 2010. [Google Scholar] [CrossRef] [PubMed]
- Spada, M.; Pagliardini, S.; Yasuda, M.; Tukel, T.; Thiagarajan, G.; Sakuraba, H.; Ponzone, A.; Desnick, R.J. High incidence of later-onset fabry disease revealed by newborn screening. Am. J. Hum. Genet. 2006, 79, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Scott, C.R.; Elliott, S.; Buroker, N.; Thomas, L.I.; Keutzer, J.; Glass, M.; Gelb, M.H.; Turecek, F. Identification of infants at risk for developing Fabry, Pompe, or mucopolysaccharidosis-I from newborn blood spots by tandem mass spectrometry. J. Pediatr. 2013, 163, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, P.V.; Campbell, C.; Klug, T.; Rogers, S.; Raburn-Miller, J.; Kiesling, J. Lysosomal storage disorder screening implementation: Findings from the first six months of full population pilot testing in missouri. J. Pediatr. 2015, 166, 172–177. [Google Scholar] [CrossRef] [PubMed]
- HGMD. Available online: http://www.Hgmd.Cf.Ac.Uk/ (accessed on 19 September 2017).
- Zimran, A.; Gelbart, T.; Westwood, B.; Grabowski, G.A.; Beutler, E. High frequency of the gaucher disease mutation at nucleotide 1226 among ashkenazi jews. Am. J. Hum. Genet. 1991, 49, 855–859. [Google Scholar] [PubMed]
- Biegstraaten, M.; Arngrimsson, R.; Barbey, F.; Boks, L.; Cecchi, F.; Deegan, P.B.; Feldt-Rasmussen, U.; Geberhiwot, T.; Germain, D.P.; Hendriksz, C.; et al. Recommendations for initiation and cessation of enzyme replacement therapy in patients with fabry disease: The european fabry working group consensus document. Orphanet J. Rare Dis. 2015, 10, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germain, D.P.; Waldek, S.; Banikazemi, M.; Bushinsky, D.A.; Charrow, J.; Desnick, R.J.; Lee, P.; Loew, T.; Vedder, A.C.; Abichandani, R.; et al. Sustained, long-term renal stabilization after 54 months of agalsidase beta therapy in patients with fabry disease. J. Am. Soc. Nephrol. 2007, 18, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Rombach, S.M.; Smid, B.E.; Bouwman, M.G.; Linthorst, G.E.; Dijkgraaf, M.G.; Hollak, C.E. Long term enzyme replacement therapy for fabry disease: Effectiveness on kidney, heart and brain. Orphanet J. Rare Dis. 2013, 8, 47. [Google Scholar] [CrossRef] [PubMed]
- Weidemann, F.; Niemann, M.; Stork, S.; Breunig, F.; Beer, M.; Sommer, C.; Herrmann, S.; Ertl, G.; Wanner, C. Long-term outcome of enzyme-replacement therapy in advanced fabry disease: Evidence for disease progression towards serious complications. J. Intern. Med. 2013, 274, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Germain, D.P.; Hughes, D.A.; Nicholls, K.; Bichet, D.G.; Giugliani, R.; Wilcox, W.R.; Feliciani, C.; Shankar, S.P.; Ezgu, F.; Amartino, H.; et al. Treatment of fabry’s disease with the pharmacologic chaperone migalastat. N. Engl. J. Med. 2016, 375, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.A.; Nicholls, K.; Shankar, S.P.; Sunder-Plassmann, G.; Koeller, D.; Nedd, K.; Vockley, G.; Hamazaki, T.; Lachmann, R.; Ohashi, T.; et al. Oral pharmacological chaperone migalastat compared with enzyme replacement therapy in fabry disease: 18-month results from the randomised phase III attract study. J. Med. Genet. 2016, 54, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Markham, A. Migalastat: First global approval. Drugs 2016, 76, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Hay Mele, B.; Citro, V.; Andreotti, G.; Cubellis, M.V. Drug repositioning can accelerate discovery of pharmacological chaperones. Orphanet J. Rare Dis. 2015, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Tierney, M.; Pottage, J.; Kessler, H.; Fischl, M.; Richman, D.; Merigan, T.; Powderly, W.; Smith, S.; Karim, A.; Sherman, J.; et al. The tolerability and pharmacokinetics of N-butyl-deoxynojirimycin in patients with advanced hiv disease (ACTG 100). The aids clinical trials group (ACTG) of the national institute of allergy and infectious diseases. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995, 10, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Butters, T.D. Pharmacotherapeutic strategies using small molecules for the treatment of glycolipid lysosomal storage disorders. Expert Opin. Pharmacother. 2007, 8, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, R.L.; D’Aquino, J.A.; Ringe, D.; Petsko, G.A. Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability. Biochemistry 2009, 48, 4816–4827. [Google Scholar] [CrossRef] [PubMed]
- Nash, R.J.; Kato, A.; Yu, C.Y.; Fleet, G.W. Iminosugars as therapeutic agents: Recent advances and promising trends. Future Med. Chem. 2011, 3, 1513–1521. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.Q.; Ishii, S.; Asano, N.; Suzuki, Y. Accelerated transport and maturation of lysosomal alpha-galactosidase a in fabry lymphoblasts by an enzyme inhibitor. Nat. Med. 1999, 5, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Hoover, J.; et al. Clinvar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016, 44, D862–D868. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. Drugbank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Andreotti, G.; Guarracino, M.R.; Cammisa, M.; Correra, A.; Cubellis, M.V. Prediction of the responsiveness to pharmacological chaperones: Lysosomal human alpha-galactosidase, a case of study. Orphanet J. Rare Dis. 2010, 5, 36. [Google Scholar] [CrossRef] [PubMed]
- Shin, M.H.; Lim, H.S. Screening methods for identifying pharmacological chaperones. Mol. Biosyst. 2017, 13, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, E.R.; Della Valle, M.C.; Wu, X.; Katz, E.; Pruthi, F.; Bond, S.; Bronfin, B.; Williams, H.; Yu, J.; Bichet, D.G.; et al. The validation of pharmacogenetics for the identification of fabry patients to be treated with migalastat. Genet. Med. 2016, 19, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Andreotti, G.; Citro, V.; Correra, A.; Cubellis, M.V. A thermodynamic assay to test pharmacological chaperones for fabry disease. Biochim. Biophys. Acta 2014, 1840, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Shabbeer, J.; Yasuda, M.; Benson, S.D.; Desnick, R.J. Fabry disease: Identification of 50 novel alpha-galactosidase a mutations causing the classic phenotype and three-dimensional structural analysis of 29 missense mutations. Hum. Genom. 2006, 2, 297–309. [Google Scholar] [CrossRef]
- Mignani, R.; Feriozzi, S.; Pisani, A.; Cioni, A.; Comotti, C.; Cossu, M.; Foschi, A.; Giudicissi, A.; Gotti, E.; Lozupone, V.A.; et al. Agalsidase therapy in patients with fabry disease on renal replacement therapy: A nationwide study in italy. Nephrol. Dial. Transplant. 2008, 23, 1628–1635. [Google Scholar] [CrossRef] [PubMed]
- Pasqualim, G.; Simon, L.; Sperb-Ludwig, F.; Burin, M.G.; Michelin-Tirelli, K.; Giugliani, R.; Matte, U. Fabry disease: A new approach for the screening of females in high-risk groups. Clin. Biochem. 2014, 47, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Von Scheidt, W.; Eng, C.M.; Fitzmaurice, T.F.; Erdmann, E.; Hubner, G.; Olsen, E.G.; Christomanou, H.; Kandolf, R.; Bishop, D.F.; Desnick, R.J. An atypical variant of fabry’s disease with manifestations confined to the myocardium. N. Engl. J. Med. 1991, 324, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Schiffmann, R.; Fuller, M.; Clarke, L.A.; Aerts, J.M. Is it fabry disease? Genet. Med. 2016, 18, 1181–1185. [Google Scholar] [CrossRef] [PubMed]
- Cammisa, M.; Correra, A.; Andreotti, G.; Cubellis, M.V. Fabry_cep: A tool to identify fabry mutations responsive to pharmacological chaperones. Orphanet J. Rare Dis. 2013, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Andreotti, G.; Citro, V.; De Crescenzo, A.; Orlando, P.; Cammisa, M.; Correra, A.; Cubellis, M.V. Therapy of fabry disease with pharmacological chaperones: From in silico predictions to in vitro tests. Orphanet J. Rare Dis. 2011, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- Lukas, J.; Giese, A.K.; Markoff, A.; Grittner, U.; Kolodny, E.; Mascher, H.; Lackner, K.J.; Meyer, W.; Wree, P.; Saviouk, V.; et al. Functional characterisation of alpha-galactosidase a mutations as a basis for a new classification system in fabry disease. PLoS Genet. 2013, 9, e1003632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cammisa, M.; Correra, A.; Andreotti, G.; Cubellis, M.V. Identification and analysis of conserved pockets on protein surfaces. BMC Bioinform. 2013, 14, S9. [Google Scholar] [CrossRef] [PubMed]
- Parenti, G.; Andria, G.; Valenzano, K.J. Pharmacological chaperone therapy: Preclinical development, clinical translation, and prospects for the treatment of lysosomal storage disorders. Mol. Ther. 2015, 23, 1138–1148. [Google Scholar] [CrossRef] [PubMed]
- Graziano, A.C.; Pannuzzo, G.; Avola, R.; Cardile, V. Chaperones as potential therapeutics for krabbe disease. J. Neurosci. Res. 2016, 94, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Henriques, B.J.; Lucas, T.G.; Gomes, C.M. Therapeutic approaches using riboflavin in mitochondrial energy metabolism disorders. Curr. Drug Targets 2016, 17, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Hole, M.; Jorge-Finnigan, A.; Underhaug, J.; Teigen, K.; Martinez, A. Pharmacological chaperones that protect tetrahydrobiopterin dependent aromatic amino acid hydroxylases through different mechanisms. Curr. Drug Targets 2016, 17, 1515–1526. [Google Scholar] [CrossRef] [PubMed]
- Banning, A.; Gulec, C.; Rouvinen, J.; Gray, S.J.; Tikkanen, R. Identification of small molecule compounds for pharmacological chaperone therapy of aspartylglucosaminuria. Sci. Rep. 2016, 6, 37583. [Google Scholar] [CrossRef] [PubMed]
- Santos-Sierra, S.; Kirchmair, J.; Perna, A.M.; Reiss, D.; Kemter, K.; Roschinger, W.; Glossmann, H.; Gersting, S.W.; Muntau, A.C.; Wolber, G.; et al. Novel pharmacological chaperones that correct phenylketonuria in mice. Hum. Mol. Genet. 2012, 21, 1877–1887. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Ohno, K.; Sakuraba, H. Fabry-database.Org: Database of the clinical phenotypes, genotypes and mutant alpha-galactosidase a structures in fabry disease. J. Hum. Genet. 2011, 56, 467–468. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cimmaruta, C.; Liguori, L.; Monticelli, M.; Andreotti, G.; Citro, V. E-Learning for Rare Diseases: An Example Using Fabry Disease. Int. J. Mol. Sci. 2017, 18, 2049. https://doi.org/10.3390/ijms18102049
Cimmaruta C, Liguori L, Monticelli M, Andreotti G, Citro V. E-Learning for Rare Diseases: An Example Using Fabry Disease. International Journal of Molecular Sciences. 2017; 18(10):2049. https://doi.org/10.3390/ijms18102049
Chicago/Turabian StyleCimmaruta, Chiara, Ludovica Liguori, Maria Monticelli, Giuseppina Andreotti, and Valentina Citro. 2017. "E-Learning for Rare Diseases: An Example Using Fabry Disease" International Journal of Molecular Sciences 18, no. 10: 2049. https://doi.org/10.3390/ijms18102049